Ruby Sedgwick
Continuous Bayesian Model Selection for Multivariate Causal Discovery
Nov 15, 2024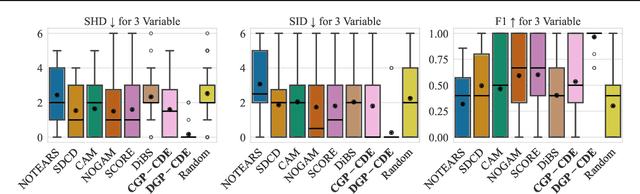
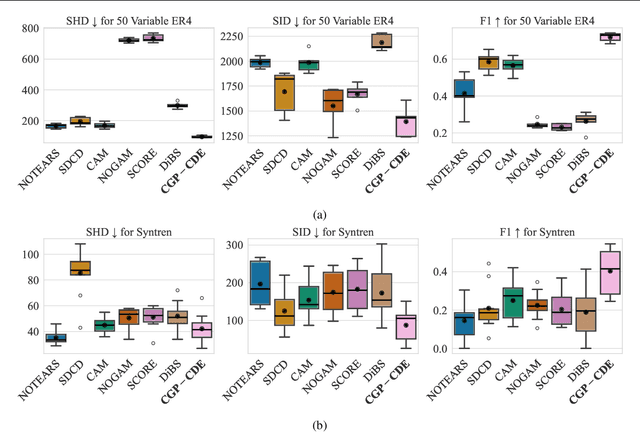
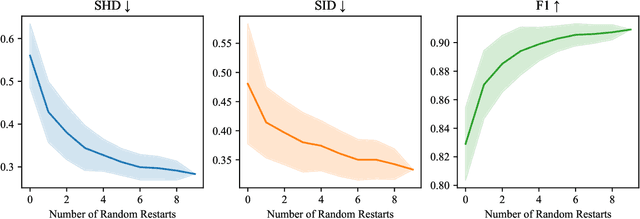
Abstract:Current causal discovery approaches require restrictive model assumptions or assume access to interventional data to ensure structure identifiability. These assumptions often do not hold in real-world applications leading to a loss of guarantees and poor accuracy in practice. Recent work has shown that, in the bivariate case, Bayesian model selection can greatly improve accuracy by exchanging restrictive modelling for more flexible assumptions, at the cost of a small probability of error. We extend the Bayesian model selection approach to the important multivariate setting by making the large discrete selection problem scalable through a continuous relaxation. We demonstrate how for our choice of Bayesian non-parametric model, the Causal Gaussian Process Conditional Density Estimator (CGP-CDE), an adjacency matrix can be constructed from the model hyperparameters. This adjacency matrix is then optimised using the marginal likelihood and an acyclicity regulariser, outputting the maximum a posteriori causal graph. We demonstrate the competitiveness of our approach on both synthetic and real-world datasets, showing it is possible to perform multivariate causal discovery without infeasible assumptions using Bayesian model selection.
Transfer Learning Bayesian Optimization to Design Competitor DNA Molecules for Use in Diagnostic Assays
Feb 27, 2024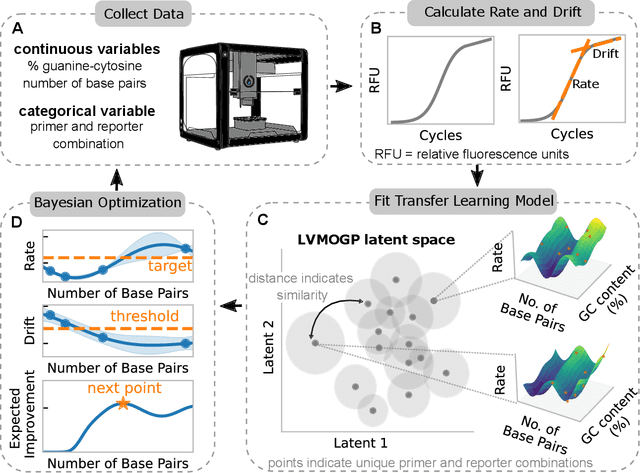
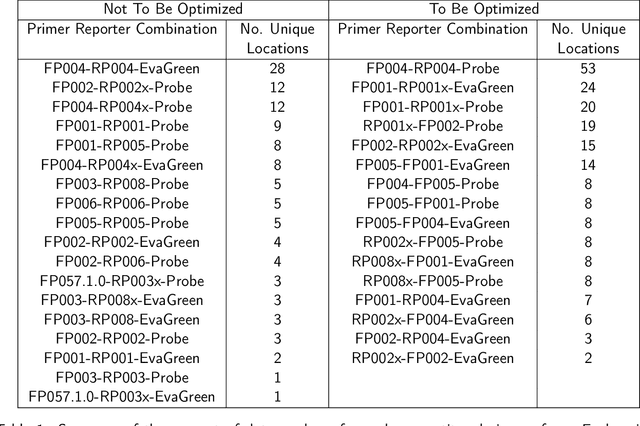
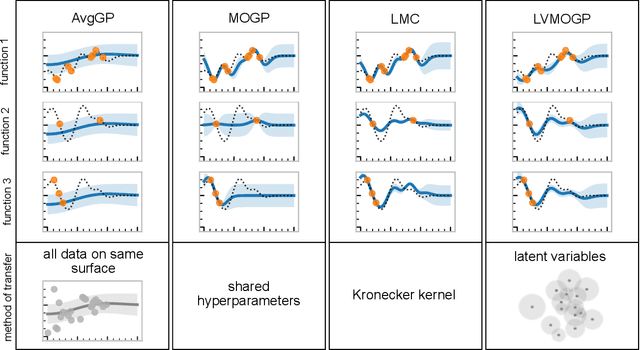
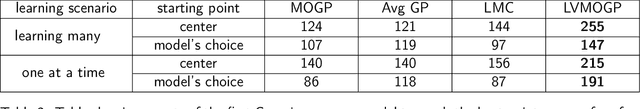
Abstract:With the rise in engineered biomolecular devices, there is an increased need for tailor-made biological sequences. Often, many similar biological sequences need to be made for a specific application meaning numerous, sometimes prohibitively expensive, lab experiments are necessary for their optimization. This paper presents a transfer learning design of experiments workflow to make this development feasible. By combining a transfer learning surrogate model with Bayesian optimization, we show how the total number of experiments can be reduced by sharing information between optimization tasks. We demonstrate the reduction in the number of experiments using data from the development of DNA competitors for use in an amplification-based diagnostic assay. We use cross-validation to compare the predictive accuracy of different transfer learning models, and then compare the performance of the models for both single objective and penalized optimization tasks.
Gaussian Processes for Monitoring Air-Quality in Kampala
Nov 28, 2023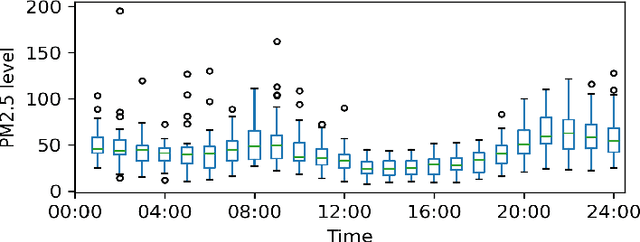
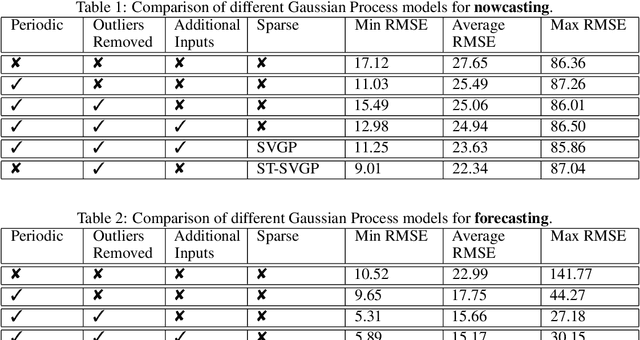
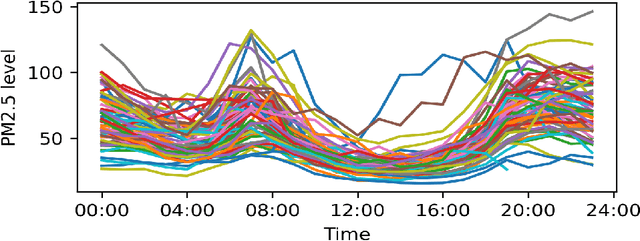
Abstract:Monitoring air pollution is of vital importance to the overall health of the population. Unfortunately, devices that can measure air quality can be expensive, and many cities in low and middle-income countries have to rely on a sparse allocation of them. In this paper, we investigate the use of Gaussian Processes for both nowcasting the current air-pollution in places where there are no sensors and forecasting the air-pollution in the future at the sensor locations. In particular, we focus on the city of Kampala in Uganda, using data from AirQo's network of sensors. We demonstrate the advantage of removing outliers, compare different kernel functions and additional inputs. We also compare two sparse approximations to allow for the large amounts of temporal data in the dataset.
Design of Experiments for Verifying Biomolecular Networks
Nov 25, 2020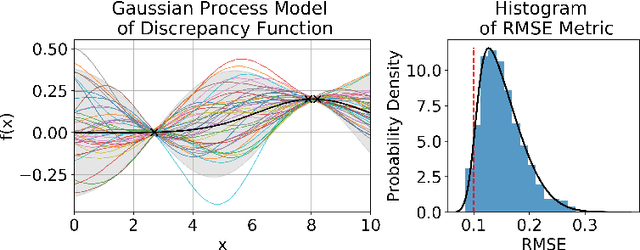
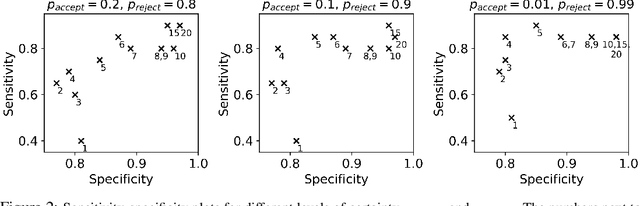
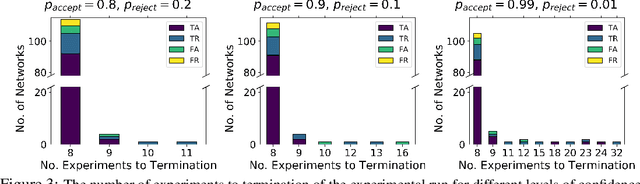
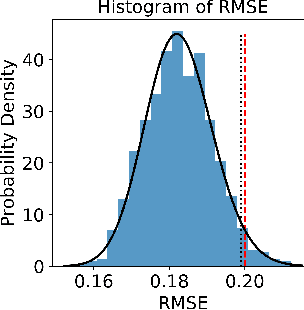
Abstract:There is a growing trend in molecular and synthetic biology of using mechanistic (non machine learning) models to design biomolecular networks. Once designed, these networks need to be validated by experimental results to ensure the theoretical network correctly models the true system. However, these experiments can be expensive and time consuming. We propose a design of experiments approach for validating these networks efficiently. Gaussian processes are used to construct a probabilistic model of the discrepancy between experimental results and the designed response, then a Bayesian optimization strategy used to select the next sample points. We compare different design criteria and develop a stopping criterion based on a metric that quantifies this discrepancy over the whole surface, and its uncertainty. We test our strategy on simulated data from computer models of biochemical processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge