Molly M. Stevens
Hyperspectral unmixing for Raman spectroscopy via physics-constrained autoencoders
Mar 07, 2024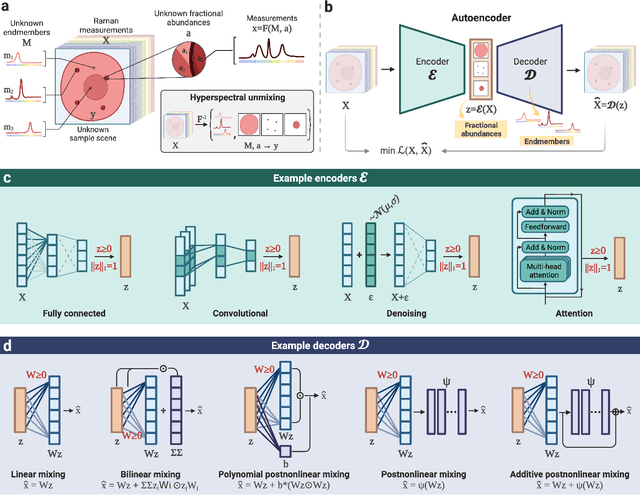
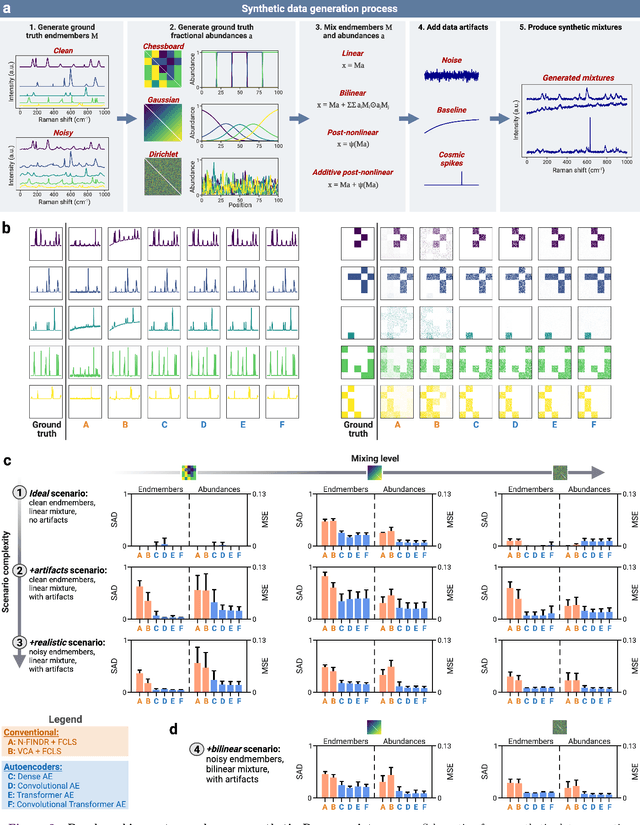
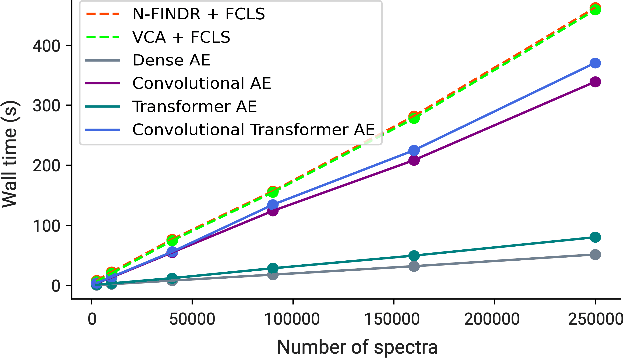
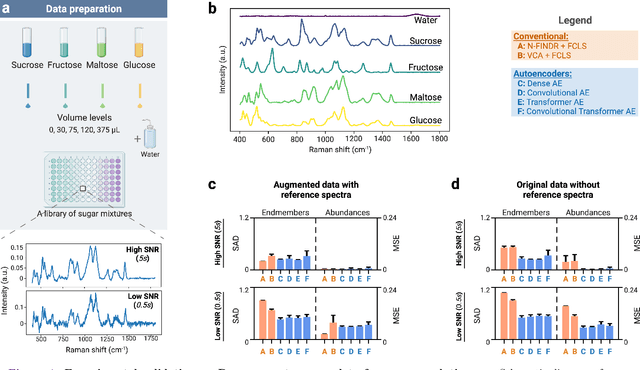
Abstract:Raman spectroscopy is widely used across scientific domains to characterize the chemical composition of samples in a non-destructive, label-free manner. Many applications entail the unmixing of signals from mixtures of molecular species to identify the individual components present and their proportions, yet conventional methods for chemometrics often struggle with complex mixture scenarios encountered in practice. Here, we develop hyperspectral unmixing algorithms based on autoencoder neural networks, and we systematically validate them using both synthetic and experimental benchmark datasets created in-house. Our results demonstrate that unmixing autoencoders provide improved accuracy, robustness and efficiency compared to standard unmixing methods. We also showcase the applicability of autoencoders to complex biological settings by showing improved biochemical characterization of volumetric Raman imaging data from a monocytic cell.
Transfer Learning Bayesian Optimization to Design Competitor DNA Molecules for Use in Diagnostic Assays
Feb 27, 2024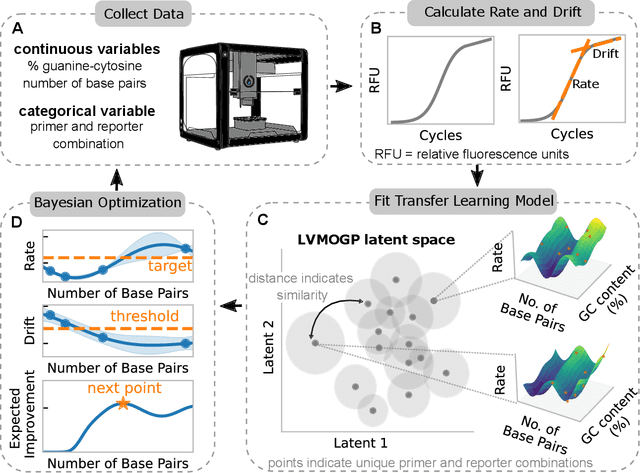
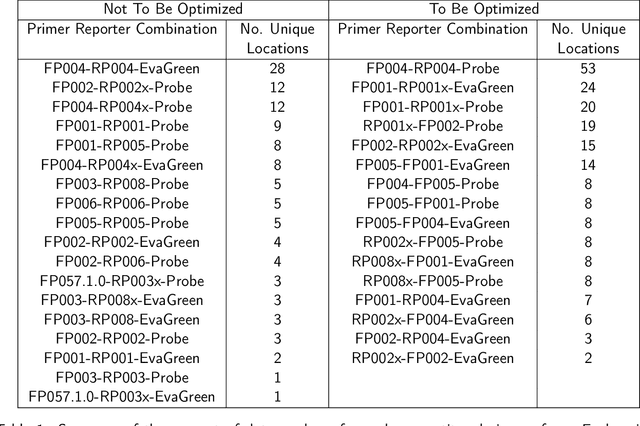
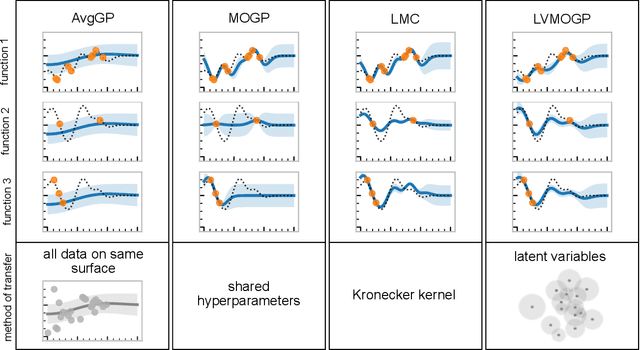
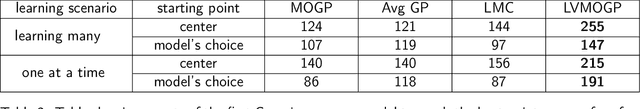
Abstract:With the rise in engineered biomolecular devices, there is an increased need for tailor-made biological sequences. Often, many similar biological sequences need to be made for a specific application meaning numerous, sometimes prohibitively expensive, lab experiments are necessary for their optimization. This paper presents a transfer learning design of experiments workflow to make this development feasible. By combining a transfer learning surrogate model with Bayesian optimization, we show how the total number of experiments can be reduced by sharing information between optimization tasks. We demonstrate the reduction in the number of experiments using data from the development of DNA competitors for use in an amplification-based diagnostic assay. We use cross-validation to compare the predictive accuracy of different transfer learning models, and then compare the performance of the models for both single objective and penalized optimization tasks.
High-throughput molecular imaging via deep learning enabled Raman spectroscopy
Sep 28, 2020


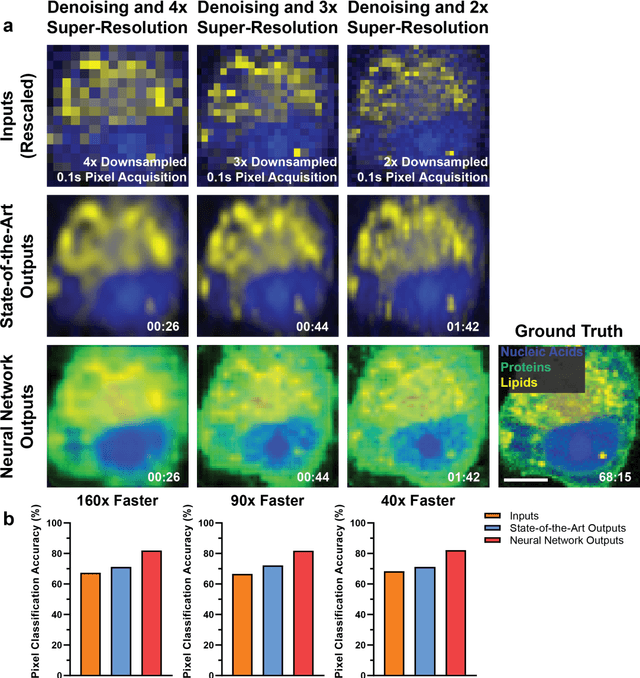
Abstract:Raman spectroscopy enables non-destructive, label-free imaging with unprecedented molecular contrast but is limited by slow data acquisition, largely preventing high-throughput imaging applications. Here, we present a comprehensive framework for higher-throughput molecular imaging via deep learning enabled Raman spectroscopy, termed DeepeR, trained on a large dataset of hyperspectral Raman images, with over 1.5 million spectra (400 hours of acquisition) in total. We firstly perform denoising and reconstruction of low signal-to-noise ratio Raman molecular signatures via deep learning, with a 9x improvement in mean squared error over state-of-the-art Raman filtering methods. Next, we develop a neural network for robust 2-4x super-resolution of hyperspectral Raman images that preserves molecular cellular information. Combining these approaches, we achieve Raman imaging speed-ups of up to 160x, enabling high resolution, high signal-to-noise ratio cellular imaging in under one minute. Finally, transfer learning is applied to extend DeepeR from cell to tissue-scale imaging. DeepeR provides a foundation that will enable a host of higher-throughput Raman spectroscopy and molecular imaging applications across biomedicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge