Conor C. Horgan
Synthetic white balancing for intra-operative hyperspectral imaging
Jul 24, 2023Abstract:Hyperspectral imaging shows promise for surgical applications to non-invasively provide spatially-resolved, spectral information. For calibration purposes, a white reference image of a highly-reflective Lambertian surface should be obtained under the same imaging conditions. Standard white references are not sterilizable, and so are unsuitable for surgical environments. We demonstrate the necessity for in situ white references and address this by proposing a novel, sterile, synthetic reference construction algorithm. The use of references obtained at different distances and lighting conditions to the subject were examined. Spectral and color reconstructions were compared with standard measurements qualitatively and quantitatively, using $\Delta E$ and normalised RMSE respectively. The algorithm forms a composite image from a video of a standard sterile ruler, whose imperfect reflectivity is compensated for. The reference is modelled as the product of independent spatial and spectral components, and a scalar factor accounting for gain, exposure, and light intensity. Evaluation of synthetic references against ideal but non-sterile references is performed using the same metrics alongside pixel-by-pixel errors. Finally, intraoperative integration is assessed though cadaveric experiments. Improper white balancing leads to increases in all quantitative and qualitative errors. Synthetic references achieve median pixel-by-pixel errors lower than 6.5% and produce similar reconstructions and errors to an ideal reference. The algorithm integrated well into surgical workflow, achieving median pixel-by-pixel errors of 4.77%, while maintaining good spectral and color reconstruction.
spectrai: A deep learning framework for spectral data
Aug 17, 2021



Abstract:Deep learning computer vision techniques have achieved many successes in recent years across numerous imaging domains. However, the application of deep learning to spectral data remains a complex task due to the need for augmentation routines, specific architectures for spectral data, and significant memory requirements. Here we present spectrai, an open-source deep learning framework designed to facilitate the training of neural networks on spectral data and enable comparison between different methods. Spectrai provides numerous built-in spectral data pre-processing and augmentation methods, neural networks for spectral data including spectral (image) denoising, spectral (image) classification, spectral image segmentation, and spectral image super-resolution. Spectrai includes both command line and graphical user interfaces (GUI) designed to guide users through model and hyperparameter decisions for a wide range of applications.
High-throughput molecular imaging via deep learning enabled Raman spectroscopy
Sep 28, 2020


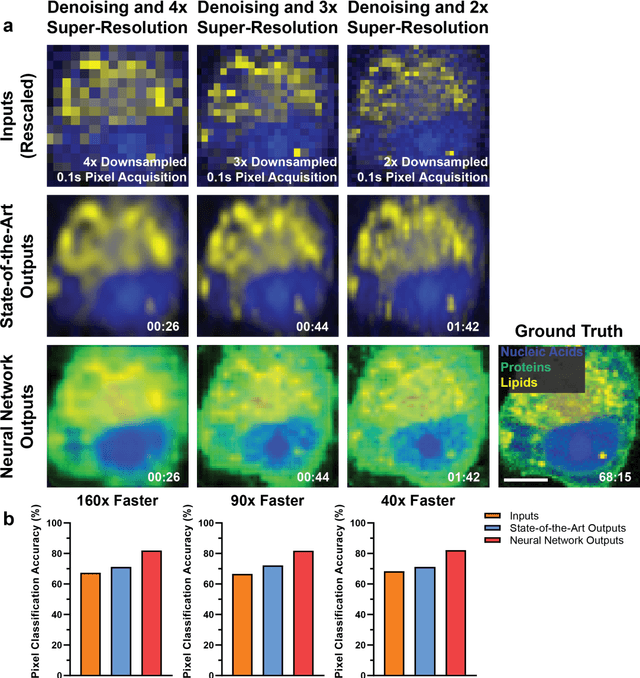
Abstract:Raman spectroscopy enables non-destructive, label-free imaging with unprecedented molecular contrast but is limited by slow data acquisition, largely preventing high-throughput imaging applications. Here, we present a comprehensive framework for higher-throughput molecular imaging via deep learning enabled Raman spectroscopy, termed DeepeR, trained on a large dataset of hyperspectral Raman images, with over 1.5 million spectra (400 hours of acquisition) in total. We firstly perform denoising and reconstruction of low signal-to-noise ratio Raman molecular signatures via deep learning, with a 9x improvement in mean squared error over state-of-the-art Raman filtering methods. Next, we develop a neural network for robust 2-4x super-resolution of hyperspectral Raman images that preserves molecular cellular information. Combining these approaches, we achieve Raman imaging speed-ups of up to 160x, enabling high resolution, high signal-to-noise ratio cellular imaging in under one minute. Finally, transfer learning is applied to extend DeepeR from cell to tissue-scale imaging. DeepeR provides a foundation that will enable a host of higher-throughput Raman spectroscopy and molecular imaging applications across biomedicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge