Avinash Kori
Object-Centric Neuro-Argumentative Learning
Jun 17, 2025Abstract:Over the last decade, as we rely more on deep learning technologies to make critical decisions, concerns regarding their safety, reliability and interpretability have emerged. We introduce a novel Neural Argumentative Learning (NAL) architecture that integrates Assumption-Based Argumentation (ABA) with deep learning for image analysis. Our architecture consists of neural and symbolic components. The former segments and encodes images into facts using object-centric learning, while the latter applies ABA learning to develop ABA frameworks enabling predictions with images. Experiments on synthetic data show that the NAL architecture can be competitive with a state-of-the-art alternative.
Decoupled Classifier-Free Guidance for Counterfactual Diffusion Models
Jun 17, 2025Abstract:Counterfactual image generation aims to simulate realistic visual outcomes under specific causal interventions. Diffusion models have recently emerged as a powerful tool for this task, combining DDIM inversion with conditional generation via classifier-free guidance (CFG). However, standard CFG applies a single global weight across all conditioning variables, which can lead to poor identity preservation and spurious attribute changes - a phenomenon known as attribute amplification. To address this, we propose Decoupled Classifier-Free Guidance (DCFG), a flexible and model-agnostic framework that introduces group-wise conditioning control. DCFG builds on an attribute-split embedding strategy that disentangles semantic inputs, enabling selective guidance on user-defined attribute groups. For counterfactual generation, we partition attributes into intervened and invariant sets based on a causal graph and apply distinct guidance to each. Experiments on CelebA-HQ, MIMIC-CXR, and EMBED show that DCFG improves intervention fidelity, mitigates unintended changes, and enhances reversibility, enabling more faithful and interpretable counterfactual image generation.
Diffusion Counterfactual Generation with Semantic Abduction
Jun 09, 2025Abstract:Counterfactual image generation presents significant challenges, including preserving identity, maintaining perceptual quality, and ensuring faithfulness to an underlying causal model. While existing auto-encoding frameworks admit semantic latent spaces which can be manipulated for causal control, they struggle with scalability and fidelity. Advancements in diffusion models present opportunities for improving counterfactual image editing, having demonstrated state-of-the-art visual quality, human-aligned perception and representation learning capabilities. Here, we present a suite of diffusion-based causal mechanisms, introducing the notions of spatial, semantic and dynamic abduction. We propose a general framework that integrates semantic representations into diffusion models through the lens of Pearlian causality to edit images via a counterfactual reasoning process. To our knowledge, this is the first work to consider high-level semantic identity preservation for diffusion counterfactuals and to demonstrate how semantic control enables principled trade-offs between faithful causal control and identity preservation.
* Proceedings of the 42nd International Conference on Machine Learning, Vancouver, Canada
Identifiable Object Representations under Spatial Ambiguities
Jun 09, 2025Abstract:Modular object-centric representations are essential for *human-like reasoning* but are challenging to obtain under spatial ambiguities, *e.g. due to occlusions and view ambiguities*. However, addressing challenges presents both theoretical and practical difficulties. We introduce a novel multi-view probabilistic approach that aggregates view-specific slots to capture *invariant content* information while simultaneously learning disentangled global *viewpoint-level* information. Unlike prior single-view methods, our approach resolves spatial ambiguities, provides theoretical guarantees for identifiability, and requires *no viewpoint annotations*. Extensive experiments on standard benchmarks and novel complex datasets validate our method's robustness and scalability.
Free Argumentative Exchanges for Explaining Image Classifiers
Feb 18, 2025Abstract:Deep learning models are powerful image classifiers but their opacity hinders their trustworthiness. Explanation methods for capturing the reasoning process within these classifiers faithfully and in a clear manner are scarce, due to their sheer complexity and size. We provide a solution for this problem by defining a novel method for explaining the outputs of image classifiers with debates between two agents, each arguing for a particular class. We obtain these debates as concrete instances of Free Argumentative eXchanges (FAXs), a novel argumentation-based multi-agent framework allowing agents to internalise opinions by other agents differently than originally stated. We define two metrics (consensus and persuasion rate) to assess the usefulness of FAXs as argumentative explanations for image classifiers. We then conduct a number of empirical experiments showing that FAXs perform well along these metrics as well as being more faithful to the image classifiers than conventional, non-argumentative explanation methods. All our implementations can be found at https://github.com/koriavinash1/FAX.
Explainable Reinforcement Learning for Formula One Race Strategy
Jan 07, 2025



Abstract:In Formula One, teams compete to develop their cars and achieve the highest possible finishing position in each race. During a race, however, teams are unable to alter the car, so they must improve their cars' finishing positions via race strategy, i.e. optimising their selection of which tyre compounds to put on the car and when to do so. In this work, we introduce a reinforcement learning model, RSRL (Race Strategy Reinforcement Learning), to control race strategies in simulations, offering a faster alternative to the industry standard of hard-coded and Monte Carlo-based race strategies. Controlling cars with a pace equating to an expected finishing position of P5.5 (where P1 represents first place and P20 is last place), RSRL achieves an average finishing position of P5.33 on our test race, the 2023 Bahrain Grand Prix, outperforming the best baseline of P5.63. We then demonstrate, in a generalisability study, how performance for one track or multiple tracks can be prioritised via training. Further, we supplement model predictions with feature importance, decision tree-based surrogate models, and decision tree counterfactuals towards improving user trust in the model. Finally, we provide illustrations which exemplify our approach in real-world situations, drawing parallels between simulations and reality.
Continuous Bayesian Model Selection for Multivariate Causal Discovery
Nov 15, 2024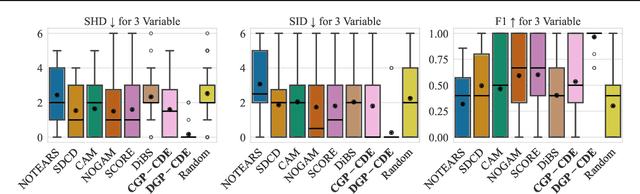
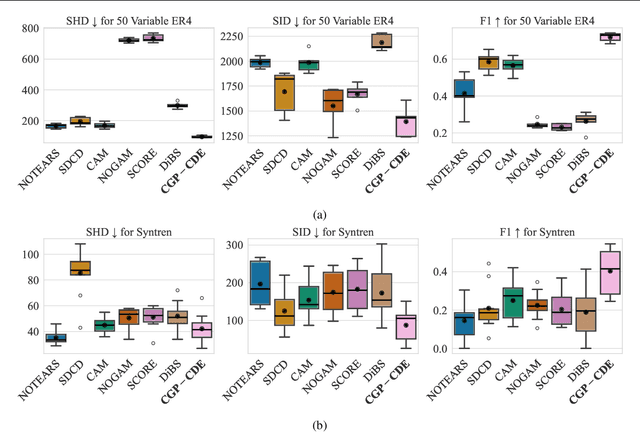
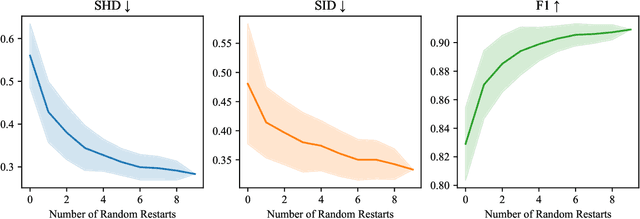
Abstract:Current causal discovery approaches require restrictive model assumptions or assume access to interventional data to ensure structure identifiability. These assumptions often do not hold in real-world applications leading to a loss of guarantees and poor accuracy in practice. Recent work has shown that, in the bivariate case, Bayesian model selection can greatly improve accuracy by exchanging restrictive modelling for more flexible assumptions, at the cost of a small probability of error. We extend the Bayesian model selection approach to the important multivariate setting by making the large discrete selection problem scalable through a continuous relaxation. We demonstrate how for our choice of Bayesian non-parametric model, the Causal Gaussian Process Conditional Density Estimator (CGP-CDE), an adjacency matrix can be constructed from the model hyperparameters. This adjacency matrix is then optimised using the marginal likelihood and an acyclicity regulariser, outputting the maximum a posteriori causal graph. We demonstrate the competitiveness of our approach on both synthetic and real-world datasets, showing it is possible to perform multivariate causal discovery without infeasible assumptions using Bayesian model selection.
Identifiable Object-Centric Representation Learning via Probabilistic Slot Attention
Jun 11, 2024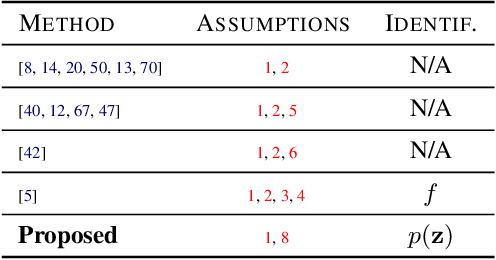
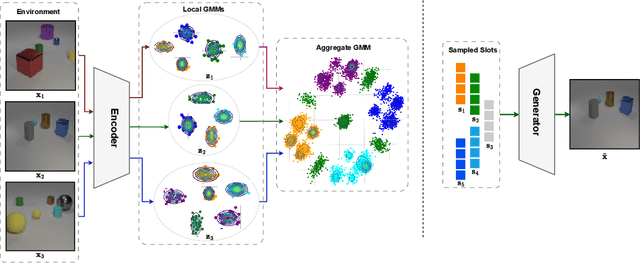
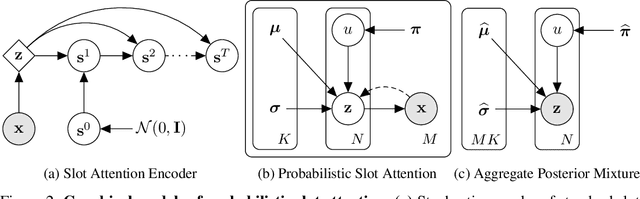
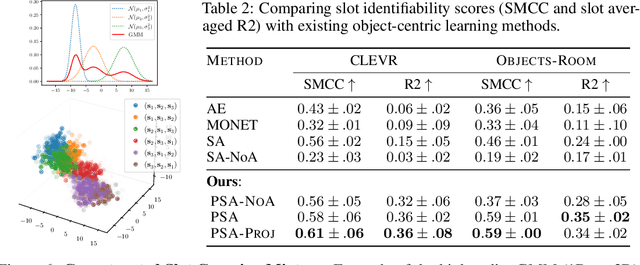
Abstract:Learning modular object-centric representations is crucial for systematic generalization. Existing methods show promising object-binding capabilities empirically, but theoretical identifiability guarantees remain relatively underdeveloped. Understanding when object-centric representations can theoretically be identified is crucial for scaling slot-based methods to high-dimensional images with correctness guarantees. To that end, we propose a probabilistic slot-attention algorithm that imposes an aggregate mixture prior over object-centric slot representations, thereby providing slot identifiability guarantees without supervision, up to an equivalence relation. We provide empirical verification of our theoretical identifiability result using both simple 2-dimensional data and high-resolution imaging datasets.
Unsupervised Conditional Slot Attention for Object Centric Learning
Jul 18, 2023



Abstract:Extracting object-level representations for downstream reasoning tasks is an emerging area in AI. Learning object-centric representations in an unsupervised setting presents multiple challenges, a key one being binding an arbitrary number of object instances to a specialized object slot. Recent object-centric representation methods like Slot Attention utilize iterative attention to learn composable representations with dynamic inference level binding but fail to achieve specialized slot level binding. To address this, in this paper we propose Unsupervised Conditional Slot Attention using a novel Probabilistic Slot Dictionary (PSD). We define PSD with (i) abstract object-level property vectors as key and (ii) parametric Gaussian distribution as its corresponding value. We demonstrate the benefits of the learnt specific object-level conditioning distributions in multiple downstream tasks, namely object discovery, compositional scene generation, and compositional visual reasoning. We show that our method provides scene composition capabilities and a significant boost in a few shot adaptability tasks of compositional visual reasoning, while performing similarly or better than slot attention in object discovery tasks
A Causal Ordering Prior for Unsupervised Representation Learning
Jul 11, 2023



Abstract:Unsupervised representation learning with variational inference relies heavily on independence assumptions over latent variables. Causal representation learning (CRL), however, argues that factors of variation in a dataset are, in fact, causally related. Allowing latent variables to be correlated, as a consequence of causal relationships, is more realistic and generalisable. So far, provably identifiable methods rely on: auxiliary information, weak labels, and interventional or even counterfactual data. Inspired by causal discovery with functional causal models, we propose a fully unsupervised representation learning method that considers a data generation process with a latent additive noise model (ANM). We encourage the latent space to follow a causal ordering via loss function based on the Hessian of the latent distribution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge