Pedro Sanchez
CheXGenBench: A Unified Benchmark For Fidelity, Privacy and Utility of Synthetic Chest Radiographs
May 15, 2025Abstract:We introduce CheXGenBench, a rigorous and multifaceted evaluation framework for synthetic chest radiograph generation that simultaneously assesses fidelity, privacy risks, and clinical utility across state-of-the-art text-to-image generative models. Despite rapid advancements in generative AI for real-world imagery, medical domain evaluations have been hindered by methodological inconsistencies, outdated architectural comparisons, and disconnected assessment criteria that rarely address the practical clinical value of synthetic samples. CheXGenBench overcomes these limitations through standardised data partitioning and a unified evaluation protocol comprising over 20 quantitative metrics that systematically analyse generation quality, potential privacy vulnerabilities, and downstream clinical applicability across 11 leading text-to-image architectures. Our results reveal critical inefficiencies in the existing evaluation protocols, particularly in assessing generative fidelity, leading to inconsistent and uninformative comparisons. Our framework establishes a standardised benchmark for the medical AI community, enabling objective and reproducible comparisons while facilitating seamless integration of both existing and future generative models. Additionally, we release a high-quality, synthetic dataset, SynthCheX-75K, comprising 75K radiographs generated by the top-performing model (Sana 0.6B) in our benchmark to support further research in this critical domain. Through CheXGenBench, we establish a new state-of-the-art and release our framework, models, and SynthCheX-75K dataset at https://raman1121.github.io/CheXGenBench/
Capacity Control is an Effective Memorization Mitigation Mechanism in Text-Conditional Diffusion Models
Oct 29, 2024

Abstract:In this work, we present compelling evidence that controlling model capacity during fine-tuning can effectively mitigate memorization in diffusion models. Specifically, we demonstrate that adopting Parameter-Efficient Fine-Tuning (PEFT) within the pre-train fine-tune paradigm significantly reduces memorization compared to traditional full fine-tuning approaches. Our experiments utilize the MIMIC dataset, which comprises image-text pairs of chest X-rays and their corresponding reports. The results, evaluated through a range of memorization and generation quality metrics, indicate that PEFT not only diminishes memorization but also enhances downstream generation quality. Additionally, PEFT methods can be seamlessly combined with existing memorization mitigation techniques for further improvement. The code for our experiments is available at: https://github.com/Raman1121/Diffusion_Memorization_HPO
MemControl: Mitigating Memorization in Medical Diffusion Models via Automated Parameter Selection
May 29, 2024



Abstract:Diffusion models show a remarkable ability in generating images that closely mirror the training distribution. However, these models are prone to training data memorization, leading to significant privacy, ethical, and legal concerns, particularly in sensitive fields such as medical imaging. We hypothesize that memorization is driven by the overparameterization of deep models, suggesting that regularizing model capacity during fine-tuning could be an effective mitigation strategy. Parameter-efficient fine-tuning (PEFT) methods offer a promising approach to capacity control by selectively updating specific parameters. However, finding the optimal subset of learnable parameters that balances generation quality and memorization remains elusive. To address this challenge, we propose a bi-level optimization framework that guides automated parameter selection by utilizing memorization and generation quality metrics as rewards. Our framework successfully identifies the optimal parameter set to be updated to satisfy the generation-memorization tradeoff. We perform our experiments for the specific task of medical image generation and outperform existing state-of-the-art training-time mitigation strategies by fine-tuning as few as 0.019% of model parameters. Furthermore, we show that the strategies learned through our framework are transferable across different datasets and domains. Our proposed framework is scalable to large datasets and agnostic to the choice of reward functions. Finally, we show that our framework can be combined with existing approaches for further memorization mitigation.
Zero-Shot Medical Phrase Grounding with Off-the-shelf Diffusion Models
Apr 19, 2024



Abstract:Localizing the exact pathological regions in a given medical scan is an important imaging problem that requires a large amount of bounding box ground truth annotations to be accurately solved. However, there exist alternative, potentially weaker, forms of supervision, such as accompanying free-text reports, which are readily available. The task of performing localization with textual guidance is commonly referred to as phrase grounding. In this work, we use a publicly available Foundation Model, namely the Latent Diffusion Model, to solve this challenging task. This choice is supported by the fact that the Latent Diffusion Model, despite being generative in nature, contains mechanisms (cross-attention) that implicitly align visual and textual features, thus leading to intermediate representations that are suitable for the task at hand. In addition, we aim to perform this task in a zero-shot manner, i.e., without any further training on target data, meaning that the model's weights remain frozen. To this end, we devise strategies to select features and also refine them via post-processing without extra learnable parameters. We compare our proposed method with state-of-the-art approaches which explicitly enforce image-text alignment in a joint embedding space via contrastive learning. Results on a popular chest X-ray benchmark indicate that our method is competitive wih SOTA on different types of pathology, and even outperforms them on average in terms of two metrics (mean IoU and AUC-ROC). Source code will be released upon acceptance.
Benchmarking Counterfactual Image Generation
Mar 29, 2024



Abstract:Counterfactual image generation is pivotal for understanding the causal relations of variables, with applications in interpretability and generation of unbiased synthetic data. However, evaluating image generation is a long-standing challenge in itself. The need to evaluate counterfactual generation compounds on this challenge, precisely because counterfactuals, by definition, are hypothetical scenarios without observable ground truths. In this paper, we present a novel comprehensive framework aimed at benchmarking counterfactual image generation methods. We incorporate metrics that focus on evaluating diverse aspects of counterfactuals, such as composition, effectiveness, minimality of interventions, and image realism. We assess the performance of three distinct conditional image generation model types, based on the Structural Causal Model paradigm. Our work is accompanied by a user-friendly Python package which allows to further evaluate and benchmark existing and future counterfactual image generation methods. Our framework is extendable to additional SCM and other causal methods, generative models, and datasets.
Group Distributionally Robust Knowledge Distillation
Nov 01, 2023Abstract:Knowledge distillation enables fast and effective transfer of features learned from a bigger model to a smaller one. However, distillation objectives are susceptible to sub-population shifts, a common scenario in medical imaging analysis which refers to groups/domains of data that are underrepresented in the training set. For instance, training models on health data acquired from multiple scanners or hospitals can yield subpar performance for minority groups. In this paper, inspired by distributionally robust optimization (DRO) techniques, we address this shortcoming by proposing a group-aware distillation loss. During optimization, a set of weights is updated based on the per-group losses at a given iteration. This way, our method can dynamically focus on groups that have low performance during training. We empirically validate our method, GroupDistil on two benchmark datasets (natural images and cardiac MRIs) and show consistent improvement in terms of worst-group accuracy.
Generative AI for Medical Imaging: extending the MONAI Framework
Jul 27, 2023Abstract:Recent advances in generative AI have brought incredible breakthroughs in several areas, including medical imaging. These generative models have tremendous potential not only to help safely share medical data via synthetic datasets but also to perform an array of diverse applications, such as anomaly detection, image-to-image translation, denoising, and MRI reconstruction. However, due to the complexity of these models, their implementation and reproducibility can be difficult. This complexity can hinder progress, act as a use barrier, and dissuade the comparison of new methods with existing works. In this study, we present MONAI Generative Models, a freely available open-source platform that allows researchers and developers to easily train, evaluate, and deploy generative models and related applications. Our platform reproduces state-of-art studies in a standardised way involving different architectures (such as diffusion models, autoregressive transformers, and GANs), and provides pre-trained models for the community. We have implemented these models in a generalisable fashion, illustrating that their results can be extended to 2D or 3D scenarios, including medical images with different modalities (like CT, MRI, and X-Ray data) and from different anatomical areas. Finally, we adopt a modular and extensible approach, ensuring long-term maintainability and the extension of current applications for future features.
A Causal Ordering Prior for Unsupervised Representation Learning
Jul 11, 2023



Abstract:Unsupervised representation learning with variational inference relies heavily on independence assumptions over latent variables. Causal representation learning (CRL), however, argues that factors of variation in a dataset are, in fact, causally related. Allowing latent variables to be correlated, as a consequence of causal relationships, is more realistic and generalisable. So far, provably identifiable methods rely on: auxiliary information, weak labels, and interventional or even counterfactual data. Inspired by causal discovery with functional causal models, we propose a fully unsupervised representation learning method that considers a data generation process with a latent additive noise model (ANM). We encourage the latent space to follow a causal ordering via loss function based on the Hessian of the latent distribution.
Compositionally Equivariant Representation Learning
Jun 17, 2023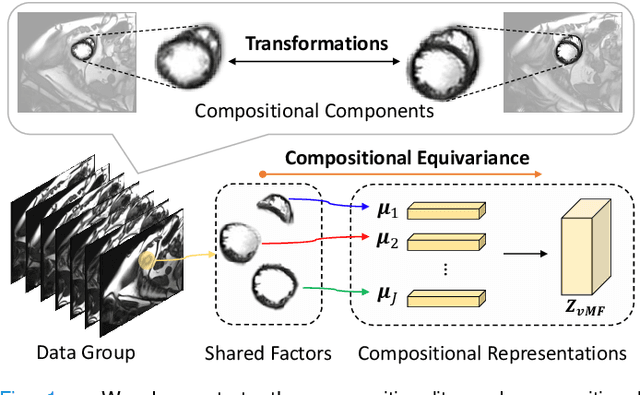
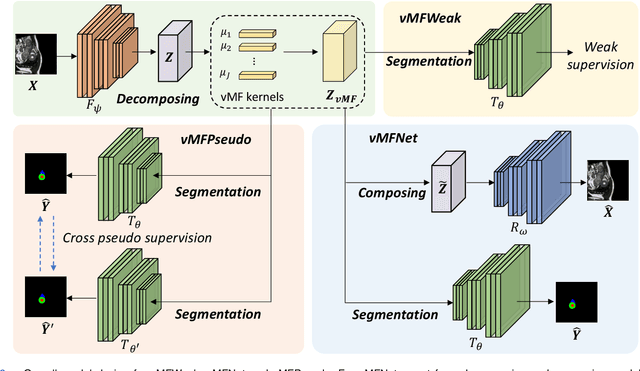
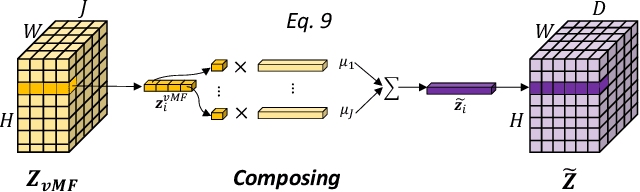
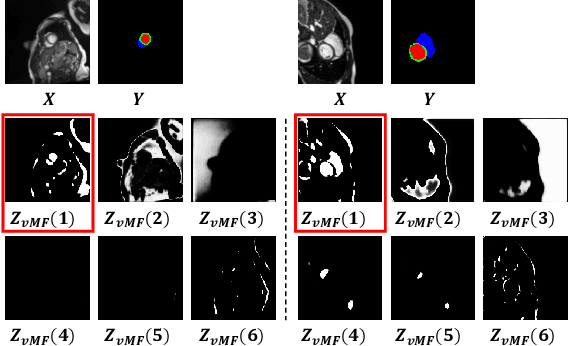
Abstract:Deep learning models often need sufficient supervision (i.e. labelled data) in order to be trained effectively. By contrast, humans can swiftly learn to identify important anatomy in medical images like MRI and CT scans, with minimal guidance. This recognition capability easily generalises to new images from different medical facilities and to new tasks in different settings. This rapid and generalisable learning ability is largely due to the compositional structure of image patterns in the human brain, which are not well represented in current medical models. In this paper, we study the utilisation of compositionality in learning more interpretable and generalisable representations for medical image segmentation. Overall, we propose that the underlying generative factors that are used to generate the medical images satisfy compositional equivariance property, where each factor is compositional (e.g. corresponds to the structures in human anatomy) and also equivariant to the task. Hence, a good representation that approximates well the ground truth factor has to be compositionally equivariant. By modelling the compositional representations with learnable von-Mises-Fisher (vMF) kernels, we explore how different design and learning biases can be used to enforce the representations to be more compositionally equivariant under un-, weakly-, and semi-supervised settings. Extensive results show that our methods achieve the best performance over several strong baselines on the task of semi-supervised domain-generalised medical image segmentation. Code will be made publicly available upon acceptance at https://github.com/vios-s.
Privacy Distillation: Reducing Re-identification Risk of Multimodal Diffusion Models
Jun 02, 2023



Abstract:Knowledge distillation in neural networks refers to compressing a large model or dataset into a smaller version of itself. We introduce Privacy Distillation, a framework that allows a text-to-image generative model to teach another model without exposing it to identifiable data. Here, we are interested in the privacy issue faced by a data provider who wishes to share their data via a multimodal generative model. A question that immediately arises is ``How can a data provider ensure that the generative model is not leaking identifiable information about a patient?''. Our solution consists of (1) training a first diffusion model on real data (2) generating a synthetic dataset using this model and filtering it to exclude images with a re-identifiability risk (3) training a second diffusion model on the filtered synthetic data only. We showcase that datasets sampled from models trained with privacy distillation can effectively reduce re-identification risk whilst maintaining downstream performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge