Ashay Patel
Resolution Invariant Autoencoder
Mar 12, 2025Abstract:Deep learning has significantly advanced medical imaging analysis, yet variations in image resolution remain an overlooked challenge. Most methods address this by resampling images, leading to either information loss or computational inefficiencies. While solutions exist for specific tasks, no unified approach has been proposed. We introduce a resolution-invariant autoencoder that adapts spatial resizing at each layer in the network via a learned variable resizing process, replacing fixed spatial down/upsampling at the traditional factor of 2. This ensures a consistent latent space resolution, regardless of input or output resolution. Our model enables various downstream tasks to be performed on an image latent whilst maintaining performance across different resolutions, overcoming the shortfalls of traditional methods. We demonstrate its effectiveness in uncertainty-aware super-resolution, classification, and generative modelling tasks and show how our method outperforms conventional baselines with minimal performance loss across resolutions.
Generative AI for Medical Imaging: extending the MONAI Framework
Jul 27, 2023Abstract:Recent advances in generative AI have brought incredible breakthroughs in several areas, including medical imaging. These generative models have tremendous potential not only to help safely share medical data via synthetic datasets but also to perform an array of diverse applications, such as anomaly detection, image-to-image translation, denoising, and MRI reconstruction. However, due to the complexity of these models, their implementation and reproducibility can be difficult. This complexity can hinder progress, act as a use barrier, and dissuade the comparison of new methods with existing works. In this study, we present MONAI Generative Models, a freely available open-source platform that allows researchers and developers to easily train, evaluate, and deploy generative models and related applications. Our platform reproduces state-of-art studies in a standardised way involving different architectures (such as diffusion models, autoregressive transformers, and GANs), and provides pre-trained models for the community. We have implemented these models in a generalisable fashion, illustrating that their results can be extended to 2D or 3D scenarios, including medical images with different modalities (like CT, MRI, and X-Ray data) and from different anatomical areas. Finally, we adopt a modular and extensible approach, ensuring long-term maintainability and the extension of current applications for future features.
Cross Attention Transformers for Multi-modal Unsupervised Whole-Body PET Anomaly Detection
Apr 14, 2023Abstract:Cancer is a highly heterogeneous condition that can occur almost anywhere in the human body. 18F-fluorodeoxyglucose is an imaging modality commonly used to detect cancer due to its high sensitivity and clear visualisation of the pattern of metabolic activity. Nonetheless, as cancer is highly heterogeneous, it is challenging to train general-purpose discriminative cancer detection models, with data availability and disease complexity often cited as a limiting factor. Unsupervised anomaly detection models have been suggested as a putative solution. These models learn a healthy representation of tissue and detect cancer by predicting deviations from the healthy norm, which requires models capable of accurately learning long-range interactions between organs and their imaging patterns with high levels of expressivity. Such characteristics are suitably satisfied by transformers, which have been shown to generate state-of-the-art results in unsupervised anomaly detection by training on normal data. This work expands upon such approaches by introducing multi-modal conditioning of the transformer via cross-attention i.e. supplying anatomical reference from paired CT. Using 294 whole-body PET/CT samples, we show that our anomaly detection method is robust and capable of achieving accurate cancer localization results even in cases where normal training data is unavailable. In addition, we show the efficacy of this approach on out-of-sample data showcasing the generalizability of this approach with limited training data. Lastly, we propose to combine model uncertainty with a new kernel density estimation approach, and show that it provides clinically and statistically significant improvements when compared to the classic residual-based anomaly maps. Overall, a superior performance is demonstrated against leading state-of-the-art alternatives, drawing attention to the potential of these approaches.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:006
PIPPI2021: An Approach to Automated Diagnosis and Texture Analysis of the Fetal Liver & Placenta in Fetal Growth Restriction
Nov 01, 2022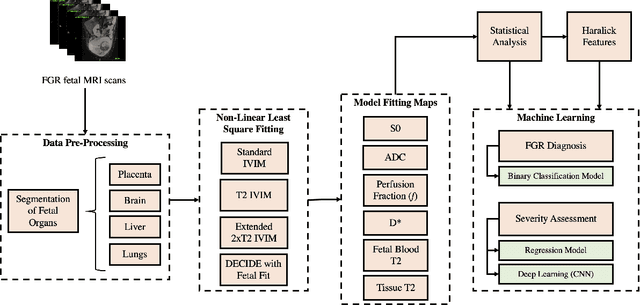
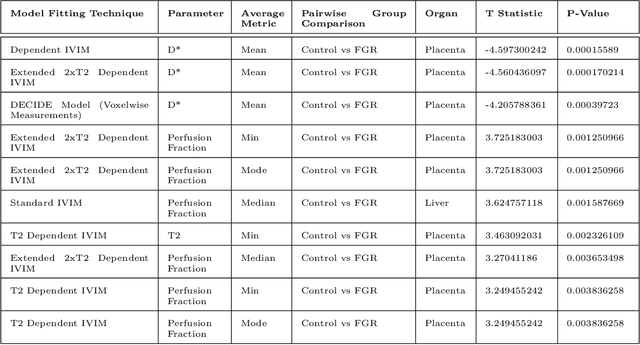
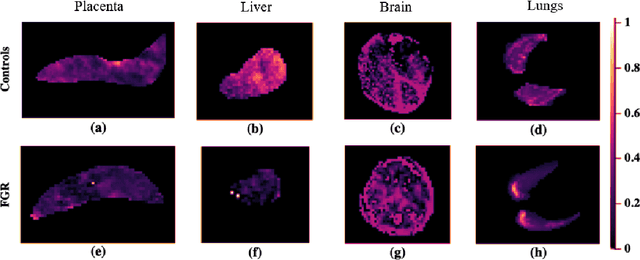
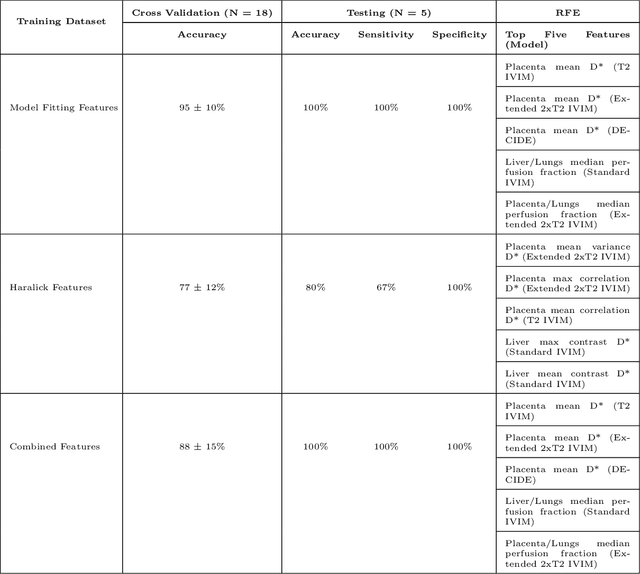
Abstract:Fetal growth restriction (FGR) is a prevalent pregnancy condition characterised by failure of the fetus to reach its genetically predetermined growth potential. We explore the application of model fitting techniques, linear regression machine learning models, deep learning regression, and Haralick textured features from multi-contrast MRI for multi-fetal organ analysis of FGR. We employed T2 relaxometry and diffusion-weighted MRI datasets (using a combined T2-diffusion scan) for 12 normally grown and 12 FGR gestational age (GA) matched pregnancies. We applied the Intravoxel Incoherent Motion Model and novel multi-compartment models for MRI fetal analysis, which exhibit potential to provide a multi-organ FGR assessment, overcoming the limitations of empirical indicators - such as abnormal artery Doppler findings - to evaluate placental dysfunction. The placenta and fetal liver presented key differentiators between FGR and normal controls (decreased perfusion, abnormal fetal blood motion and reduced fetal blood oxygenation. This may be associated with the preferential shunting of the fetal blood towards the fetal brain. These features were further explored to determine their role in assessing FGR severity, by employing simple machine learning models to predict FGR diagnosis (100\% accuracy in test data, n=5), GA at delivery, time from MRI scan to delivery, and baby weight. Moreover, we explored the use of deep learning to regress the latter three variables. Image texture analysis of the fetal organs demonstrated prominent textural variations in the placental perfusion fractions maps between the groups (p$<$0.0009), and spatial differences in the incoherent fetal capillary blood motion in the liver (p$<$0.009). This research serves as a proof-of-concept, investigating the effect of FGR on fetal organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge