Athanasios Vlontzos
Causally Steered Diffusion for Automated Video Counterfactual Generation
Jun 17, 2025Abstract:Adapting text-to-image (T2I) latent diffusion models for video editing has shown strong visual fidelity and controllability, but challenges remain in maintaining causal relationships in video content. Edits affecting causally dependent attributes risk generating unrealistic or misleading outcomes if these relationships are ignored. In this work, we propose a causally faithful framework for counterfactual video generation, guided by a vision-language model (VLM). Our method is agnostic to the underlying video editing system and does not require access to its internal mechanisms or finetuning. Instead, we guide the generation by optimizing text prompts based on an assumed causal graph, addressing the challenge of latent space control in LDMs. We evaluate our approach using standard video quality metrics and counterfactual-specific criteria, such as causal effectiveness and minimality. Our results demonstrate that causally faithful video counterfactuals can be effectively generated within the learned distribution of LDMs through prompt-based causal steering. With its compatibility with any black-box video editing system, our method holds significant potential for generating realistic "what-if" video scenarios in diverse areas such as healthcare and digital media.
The Hardness of Validating Observational Studies with Experimental Data
Mar 19, 2025Abstract:Observational data is often readily available in large quantities, but can lead to biased causal effect estimates due to the presence of unobserved confounding. Recent works attempt to remove this bias by supplementing observational data with experimental data, which, when available, is typically on a smaller scale due to the time and cost involved in running a randomised controlled trial. In this work, we prove a theorem that places fundamental limits on this ``best of both worlds'' approach. Using the framework of impossible inference, we show that although it is possible to use experimental data to \emph{falsify} causal effect estimates from observational data, in general it is not possible to \emph{validate} such estimates. Our theorem proves that while experimental data can be used to detect bias in observational studies, without additional assumptions on the smoothness of the correction function, it can not be used to remove it. We provide a practical example of such an assumption, developing a novel Gaussian Process based approach to construct intervals which contain the true treatment effect with high probability, both inside and outside of the support of the experimental data. We demonstrate our methodology on both simulated and semi-synthetic datasets and make the \href{https://github.com/Jakefawkes/Obs_and_exp_data}{code available}.
Contrastive representations of high-dimensional, structured treatments
Nov 28, 2024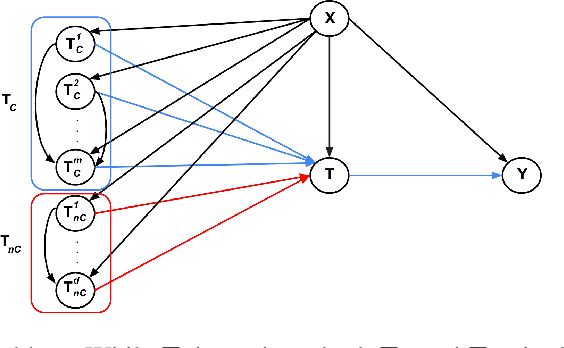



Abstract:Estimating causal effects is vital for decision making. In standard causal effect estimation, treatments are usually binary- or continuous-valued. However, in many important real-world settings, treatments can be structured, high-dimensional objects, such as text, video, or audio. This provides a challenge to traditional causal effect estimation. While leveraging the shared structure across different treatments can help generalize to unseen treatments at test time, we show in this paper that using such structure blindly can lead to biased causal effect estimation. We address this challenge by devising a novel contrastive approach to learn a representation of the high-dimensional treatments, and prove that it identifies underlying causal factors and discards non-causally relevant factors. We prove that this treatment representation leads to unbiased estimates of the causal effect, and empirically validate and benchmark our results on synthetic and real-world datasets.
Benchmarking Counterfactual Image Generation
Mar 29, 2024



Abstract:Counterfactual image generation is pivotal for understanding the causal relations of variables, with applications in interpretability and generation of unbiased synthetic data. However, evaluating image generation is a long-standing challenge in itself. The need to evaluate counterfactual generation compounds on this challenge, precisely because counterfactuals, by definition, are hypothetical scenarios without observable ground truths. In this paper, we present a novel comprehensive framework aimed at benchmarking counterfactual image generation methods. We incorporate metrics that focus on evaluating diverse aspects of counterfactuals, such as composition, effectiveness, minimality of interventions, and image realism. We assess the performance of three distinct conditional image generation model types, based on the Structural Causal Model paradigm. Our work is accompanied by a user-friendly Python package which allows to further evaluate and benchmark existing and future counterfactual image generation methods. Our framework is extendable to additional SCM and other causal methods, generative models, and datasets.
Non-parametric identifiability and sensitivity analysis of synthetic control models
Jan 18, 2023



Abstract:Quantifying cause and effect relationships is an important problem in many domains. The gold standard solution is to conduct a randomised controlled trial. However, in many situations such trials cannot be performed. In the absence of such trials, many methods have been devised to quantify the causal impact of an intervention from observational data given certain assumptions. One widely used method are synthetic control models. While identifiability of the causal estimand in such models has been obtained from a range of assumptions, it is widely and implicitly assumed that the underlying assumptions are satisfied for all time periods both pre- and post-intervention. This is a strong assumption, as synthetic control models can only be learned in pre-intervention period. In this paper we address this challenge, and prove identifiability can be obtained without the need for this assumption, by showing it follows from the principle of invariant causal mechanisms. Moreover, for the first time, we formulate and study synthetic control models in Pearl's structural causal model framework. Importantly, we provide a general framework for sensitivity analysis of synthetic control causal inference to violations of the assumptions underlying non-parametric identifiability. We end by providing an empirical demonstration of our sensitivity analysis framework on simulated and real data in the widely-used linear synthetic control framework.
Self-Supervised 3D Human Pose Estimation in Static Video Via Neural Rendering
Oct 10, 2022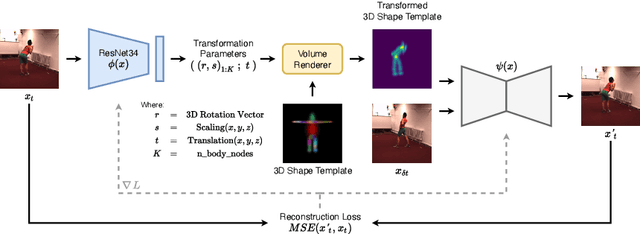
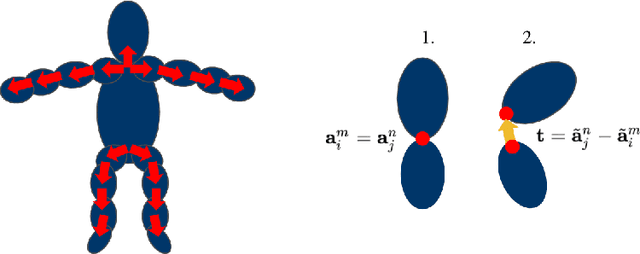
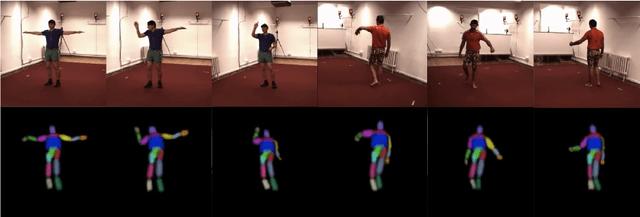
Abstract:Inferring 3D human pose from 2D images is a challenging and long-standing problem in the field of computer vision with many applications including motion capture, virtual reality, surveillance or gait analysis for sports and medicine. We present preliminary results for a method to estimate 3D pose from 2D video containing a single person and a static background without the need for any manual landmark annotations. We achieve this by formulating a simple yet effective self-supervision task: our model is required to reconstruct a random frame of a video given a frame from another timepoint and a rendered image of a transformed human shape template. Crucially for optimisation, our ray casting based rendering pipeline is fully differentiable, enabling end to end training solely based on the reconstruction task.
Adnexal Mass Segmentation with Ultrasound Data Synthesis
Sep 25, 2022Abstract:Ovarian cancer is the most lethal gynaecological malignancy. The disease is most commonly asymptomatic at its early stages and its diagnosis relies on expert evaluation of transvaginal ultrasound images. Ultrasound is the first-line imaging modality for characterising adnexal masses, it requires significant expertise and its analysis is subjective and labour-intensive, therefore open to error. Hence, automating processes to facilitate and standardise the evaluation of scans is desired in clinical practice. Using supervised learning, we have demonstrated that segmentation of adnexal masses is possible, however, prevalence and label imbalance restricts the performance on under-represented classes. To mitigate this we apply a novel pathology-specific data synthesiser. We create synthetic medical images with their corresponding ground truth segmentations by using Poisson image editing to integrate less common masses into other samples. Our approach achieves the best performance across all classes, including an improvement of up to 8% when compared with nnU-Net baseline approaches.
nnOOD: A Framework for Benchmarking Self-supervised Anomaly Localisation Methods
Sep 02, 2022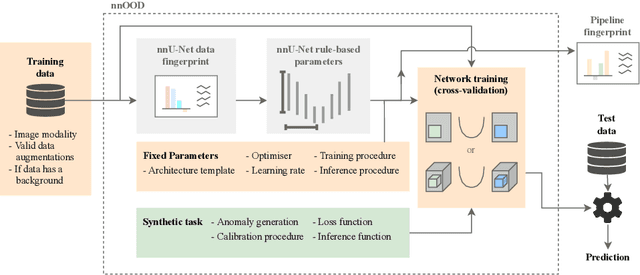
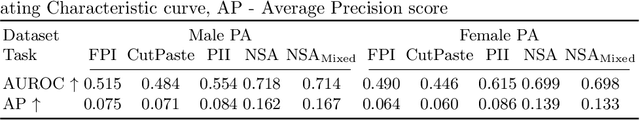
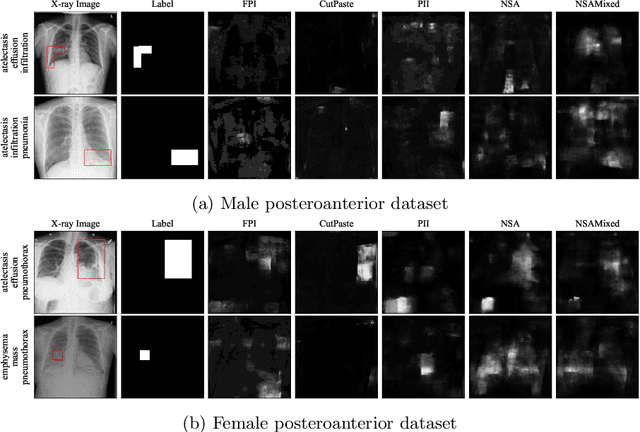
Abstract:The wide variety of in-distribution and out-of-distribution data in medical imaging makes universal anomaly detection a challenging task. Recently a number of self-supervised methods have been developed that train end-to-end models on healthy data augmented with synthetic anomalies. However, it is difficult to compare these methods as it is not clear whether gains in performance are from the task itself or the training pipeline around it. It is also difficult to assess whether a task generalises well for universal anomaly detection, as they are often only tested on a limited range of anomalies. To assist with this we have developed nnOOD, a framework that adapts nnU-Net to allow for comparison of self-supervised anomaly localisation methods. By isolating the synthetic, self-supervised task from the rest of the training process we perform a more faithful comparison of the tasks, whilst also making the workflow for evaluating over a given dataset quick and easy. Using this we have implemented the current state-of-the-art tasks and evaluated them on a challenging X-ray dataset.
A Review of Causality for Learning Algorithms in Medical Image Analysis
Jun 11, 2022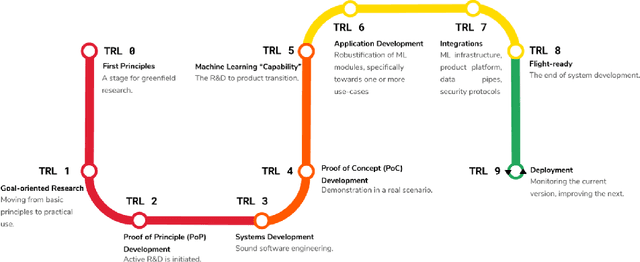
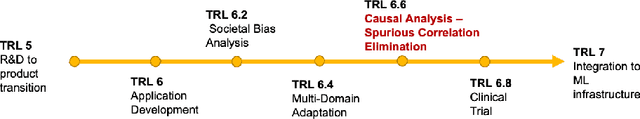
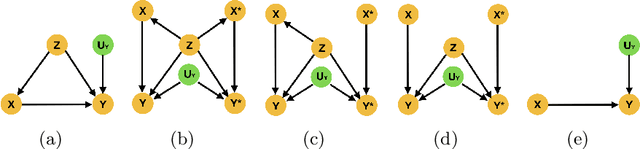
Abstract:Medical image analysis is a vibrant research area that offers doctors and medical practitioners invaluable insight and the ability to accurately diagnose and monitor disease. Machine learning provides an additional boost for this area. However, machine learning for medical image analysis is particularly vulnerable to natural biases like domain shifts that affect algorithmic performance and robustness. In this paper we analyze machine learning for medical image analysis within the framework of Technology Readiness Levels and review how causal analysis methods can fill a gap when creating robust and adaptable medical image analysis algorithms. We review methods using causality in medical imaging AI/ML and find that causal analysis has the potential to mitigate critical problems for clinical translation but that uptake and clinical downstream research has been limited so far.
Is More Data All You Need? A Causal Exploration
Jun 06, 2022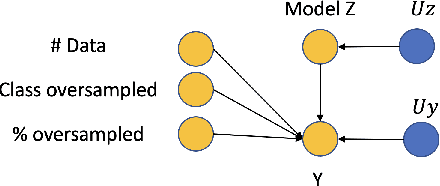
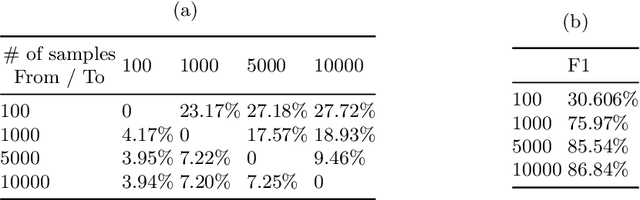
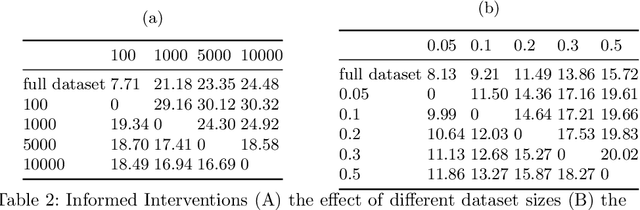
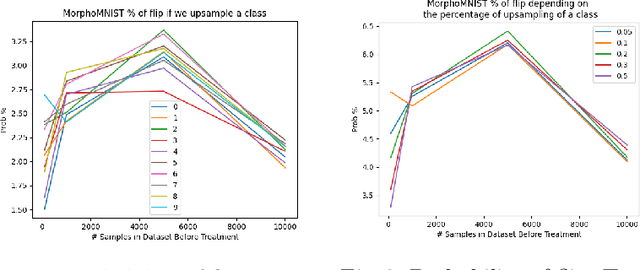
Abstract:Curating a large scale medical imaging dataset for machine learning applications is both time consuming and expensive. Balancing the workload between model development, data collection and annotations is difficult for machine learning practitioners, especially under time constraints. Causal analysis is often used in medicine and economics to gain insights about the effects of actions and policies. In this paper we explore the effect of dataset interventions on the output of image classification models. Through a causal approach we investigate the effects of the quantity and type of data we need to incorporate in a dataset to achieve better performance for specific subtasks. The main goal of this paper is to highlight the potential of causal analysis as a tool for resource optimization for developing medical imaging ML applications. We explore this concept with a synthetic dataset and an exemplary use-case for Diabetic Retinopathy image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge