Spyridon Thermos
Towards Practical Single-shot Motion Synthesis
Jun 04, 2024Abstract:Despite the recent advances in the so-called "cold start" generation from text prompts, their needs in data and computing resources, as well as the ambiguities around intellectual property and privacy concerns pose certain counterarguments for their utility. An interesting and relatively unexplored alternative has been the introduction of unconditional synthesis from a single sample, which has led to interesting generative applications. In this paper we focus on single-shot motion generation and more specifically on accelerating the training time of a Generative Adversarial Network (GAN). In particular, we tackle the challenge of GAN's equilibrium collapse when using mini-batch training by carefully annealing the weights of the loss functions that prevent mode collapse. Additionally, we perform statistical analysis in the generator and discriminator models to identify correlations between training stages and enable transfer learning. Our improved GAN achieves competitive quality and diversity on the Mixamo benchmark when compared to the original GAN architecture and a single-shot diffusion model, while being up to x6.8 faster in training time from the former and x1.75 from the latter. Finally, we demonstrate the ability of our improved GAN to mix and compose motion with a single forward pass. Project page available at https://moverseai.github.io/single-shot.
Noise-in, Bias-out: Balanced and Real-time MoCap Solving
Sep 25, 2023



Abstract:Real-time optical Motion Capture (MoCap) systems have not benefited from the advances in modern data-driven modeling. In this work we apply machine learning to solve noisy unstructured marker estimates in real-time and deliver robust marker-based MoCap even when using sparse affordable sensors. To achieve this we focus on a number of challenges related to model training, namely the sourcing of training data and their long-tailed distribution. Leveraging representation learning we design a technique for imbalanced regression that requires no additional data or labels and improves the performance of our model in rare and challenging poses. By relying on a unified representation, we show that training such a model is not bound to high-end MoCap training data acquisition, and exploit the advances in marker-less MoCap to acquire the necessary data. Finally, we take a step towards richer and affordable MoCap by adapting a body model-based inverse kinematics solution to account for measurement and inference uncertainty, further improving performance and robustness. Project page: https://moverseai.github.io/noise-tail
Compositionally Equivariant Representation Learning
Jun 17, 2023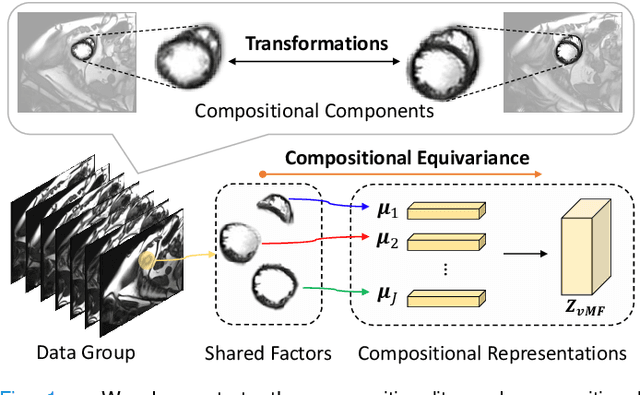
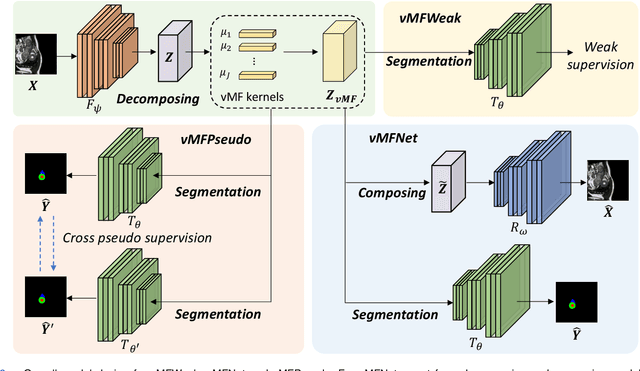
Abstract:Deep learning models often need sufficient supervision (i.e. labelled data) in order to be trained effectively. By contrast, humans can swiftly learn to identify important anatomy in medical images like MRI and CT scans, with minimal guidance. This recognition capability easily generalises to new images from different medical facilities and to new tasks in different settings. This rapid and generalisable learning ability is largely due to the compositional structure of image patterns in the human brain, which are not well represented in current medical models. In this paper, we study the utilisation of compositionality in learning more interpretable and generalisable representations for medical image segmentation. Overall, we propose that the underlying generative factors that are used to generate the medical images satisfy compositional equivariance property, where each factor is compositional (e.g. corresponds to the structures in human anatomy) and also equivariant to the task. Hence, a good representation that approximates well the ground truth factor has to be compositionally equivariant. By modelling the compositional representations with learnable von-Mises-Fisher (vMF) kernels, we explore how different design and learning biases can be used to enforce the representations to be more compositionally equivariant under un-, weakly-, and semi-supervised settings. Extensive results show that our methods achieve the best performance over several strong baselines on the task of semi-supervised domain-generalised medical image segmentation. Code will be made publicly available upon acceptance at https://github.com/vios-s.
HSIC-InfoGAN: Learning Unsupervised Disentangled Representations by Maximising Approximated Mutual Information
Aug 06, 2022Abstract:Learning disentangled representations requires either supervision or the introduction of specific model designs and learning constraints as biases. InfoGAN is a popular disentanglement framework that learns unsupervised disentangled representations by maximising the mutual information between latent representations and their corresponding generated images. Maximisation of mutual information is achieved by introducing an auxiliary network and training with a latent regression loss. In this short exploratory paper, we study the use of the Hilbert-Schmidt Independence Criterion (HSIC) to approximate mutual information between latent representation and image, termed HSIC-InfoGAN. Directly optimising the HSIC loss avoids the need for an additional auxiliary network. We qualitatively compare the level of disentanglement in each model, suggest a strategy to tune the hyperparameters of HSIC-InfoGAN, and discuss the potential of HSIC-InfoGAN for medical applications.
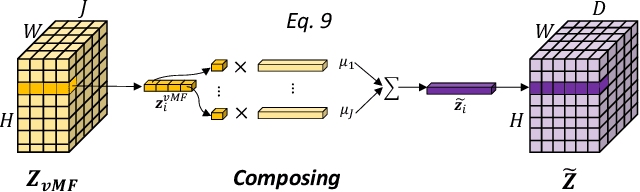
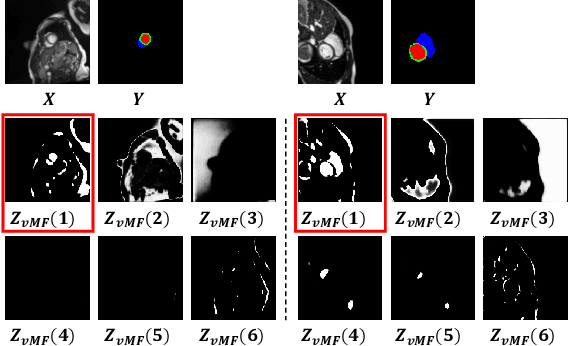
vMFNet: Compositionality Meets Domain-generalised Segmentation
Jun 29, 2022


Abstract:Training medical image segmentation models usually requires a large amount of labeled data. By contrast, humans can quickly learn to accurately recognise anatomy of interest from medical (e.g. MRI and CT) images with some limited guidance. Such recognition ability can easily generalise to new images from different clinical centres. This rapid and generalisable learning ability is mostly due to the compositional structure of image patterns in the human brain, which is less incorporated in medical image segmentation. In this paper, we model the compositional components (i.e. patterns) of human anatomy as learnable von-Mises-Fisher (vMF) kernels, which are robust to images collected from different domains (e.g. clinical centres). The image features can be decomposed to (or composed by) the components with the composing operations, i.e. the vMF likelihoods. The vMF likelihoods tell how likely each anatomical part is at each position of the image. Hence, the segmentation mask can be predicted based on the vMF likelihoods. Moreover, with a reconstruction module, unlabeled data can also be used to learn the vMF kernels and likelihoods by recombining them to reconstruct the input image. Extensive experiments show that the proposed vMFNet achieves improved generalisation performance on two benchmarks, especially when annotations are limited. Code is publicly available at: https://github.com/vios-s/vMFNet.
A Tutorial on Learning Disentangled Representations in the Imaging Domain
Aug 26, 2021



Abstract:Disentangled representation learning has been proposed as an approach to learning general representations. This can be done in the absence of, or with limited, annotations. A good general representation can be readily fine-tuned for new target tasks using modest amounts of data, or even be used directly in unseen domains achieving remarkable performance in the corresponding task. This alleviation of the data and annotation requirements offers tantalising prospects for tractable and affordable applications in computer vision and healthcare. Finally, disentangled representations can offer model explainability and can help us understand the underlying causal relations of the factors of variation, increasing their suitability for real-world deployment. In this tutorial paper, we will offer an overview of the disentangled representation learning, its building blocks and criteria, and discuss applications in computer vision and medical imaging. We conclude our tutorial by presenting the identified opportunities for the integration of recent machine learning advances into disentanglement, as well as the remaining challenges.
Controllable cardiac synthesis via disentangled anatomy arithmetic
Jul 04, 2021



Abstract:Acquiring annotated data at scale with rare diseases or conditions remains a challenge. It would be extremely useful to have a method that controllably synthesizes images that can correct such underrepresentation. Assuming a proper latent representation, the idea of a "latent vector arithmetic" could offer the means of achieving such synthesis. A proper representation must encode the fidelity of the input data, preserve invariance and equivariance, and permit arithmetic operations. Motivated by the ability to disentangle images into spatial anatomy (tensor) factors and accompanying imaging (vector) representations, we propose a framework termed "disentangled anatomy arithmetic", in which a generative model learns to combine anatomical factors of different input images such that when they are re-entangled with the desired imaging modality (e.g. MRI), plausible new cardiac images are created with the target characteristics. To encourage a realistic combination of anatomy factors after the arithmetic step, we propose a localized noise injection network that precedes the generator. Our model is used to generate realistic images, pathology labels, and segmentation masks that are used to augment the existing datasets and subsequently improve post-hoc classification and segmentation tasks. Code is publicly available at https://github.com/vios-s/DAA-GAN.
Semi-supervised Meta-learning with Disentanglement for Domain-generalised Medical Image Segmentation
Jun 24, 2021



Abstract:Generalising deep models to new data from new centres (termed here domains) remains a challenge. This is largely attributed to shifts in data statistics (domain shifts) between source and unseen domains. Recently, gradient-based meta-learning approaches where the training data are split into meta-train and meta-test sets to simulate and handle the domain shifts during training have shown improved generalisation performance. However, the current fully supervised meta-learning approaches are not scalable for medical image segmentation, where large effort is required to create pixel-wise annotations. Meanwhile, in a low data regime, the simulated domain shifts may not approximate the true domain shifts well across source and unseen domains. To address this problem, we propose a novel semi-supervised meta-learning framework with disentanglement. We explicitly model the representations related to domain shifts. Disentangling the representations and combining them to reconstruct the input image allows unlabeled data to be used to better approximate the true domain shifts for meta-learning. Hence, the model can achieve better generalisation performance, especially when there is a limited amount of labeled data. Experiments show that the proposed method is robust on different segmentation tasks and achieves state-of-the-art generalisation performance on two public benchmarks.
Metrics for Exposing the Biases of Content-Style Disentanglement
Aug 31, 2020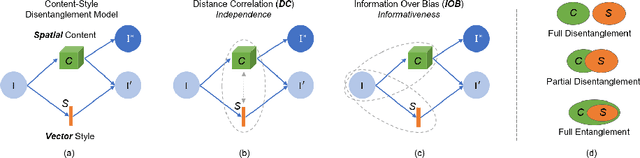
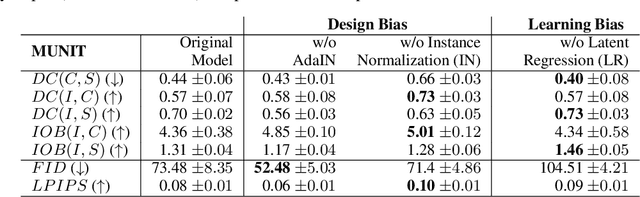
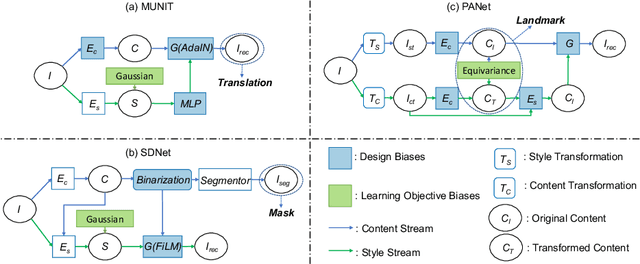

Abstract:Recent state-of-the-art semi- and un-supervised solutions for challenging computer vision tasks have used the idea of encoding image content into a spatial tensor and image appearance or "style" into a vector. These decomposed representations take advantage of equivariant properties of network design and improve performance in equivariant tasks, such as image-to-image translation. Most of these methods use the term "disentangled" for their representations and employ model design, learning objectives, and data biases to achieve good model performance. While considerable effort has been made to measure disentanglement in vector representations, currently, metrics that can characterize the degree of disentanglement between content (spatial) and style (vector) representations and the relation to task performance are lacking. In this paper, we propose metrics to measure how (un)correlated, biased, and informative the content and style representations are. In particular, we first identify key design choices and learning constraints on three popular models that employ content-style disentanglement and derive ablated versions. Then, we use our metrics to ascertain the role of each bias. Our experiments reveal a "sweet-spot" between disentanglement, task performance and latent space interpretability. The proposed metrics enable the design of better models and the selection of models that achieve the desired performance and disentanglement. Our metrics library is available at https://github.com/TsaftarisCollaboratory/CSDisentanglement_Metrics_Library.
Disentangled Representations for Domain-generalized Cardiac Segmentation
Aug 26, 2020


Abstract:Robust cardiac image segmentation is still an open challenge due to the inability of the existing methods to achieve satisfactory performance on unseen data of different domains. Since the acquisition and annotation of medical data are costly and time-consuming, recent work focuses on domain adaptation and generalization to bridge the gap between data from different populations and scanners. In this paper, we propose two data augmentation methods that focus on improving the domain adaptation and generalization abilities of state-to-the-art cardiac segmentation models. In particular, our "Resolution Augmentation" method generates more diverse data by rescaling images to different resolutions within a range spanning different scanner protocols. Subsequently, our "Factor-based Augmentation" method generates more diverse data by projecting the original samples onto disentangled latent spaces, and combining the learned anatomy and modality factors from different domains. Our extensive experiments demonstrate the importance of efficient adaptation between seen and unseen domains, as well as model generalization ability, to robust cardiac image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge