Design of Experiments for Verifying Biomolecular Networks
Paper and Code
Nov 25, 2020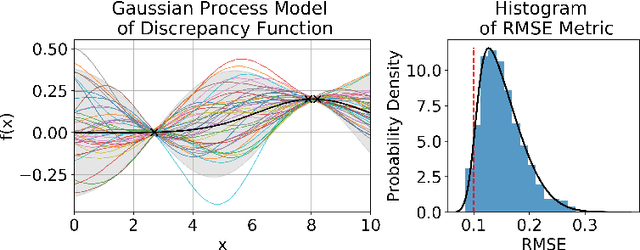
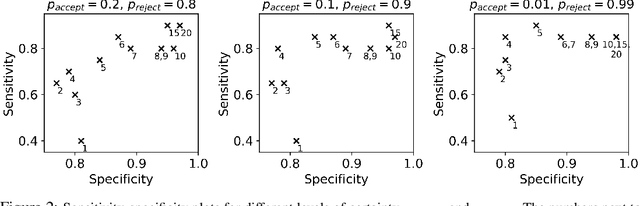
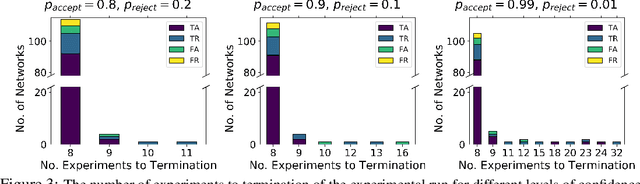
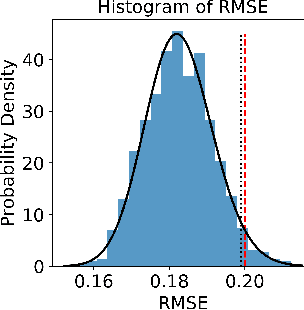
There is a growing trend in molecular and synthetic biology of using mechanistic (non machine learning) models to design biomolecular networks. Once designed, these networks need to be validated by experimental results to ensure the theoretical network correctly models the true system. However, these experiments can be expensive and time consuming. We propose a design of experiments approach for validating these networks efficiently. Gaussian processes are used to construct a probabilistic model of the discrepancy between experimental results and the designed response, then a Bayesian optimization strategy used to select the next sample points. We compare different design criteria and develop a stopping criterion based on a metric that quantifies this discrepancy over the whole surface, and its uncertainty. We test our strategy on simulated data from computer models of biochemical processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge