Stefan Winzeck
Risk of Bias in Chest X-ray Foundation Models
Sep 07, 2022

Abstract:Foundation models are considered a breakthrough in all applications of AI, promising robust and reusable mechanisms for feature extraction, alleviating the need for large amounts of high quality training data for task-specific prediction models. However, foundation models may potentially encode and even reinforce existing biases present in historic datasets. Given the limited ability to scrutinize foundation models, it remains unclear whether the opportunities outweigh the risks in safety critical applications such as clinical decision making. In our statistical bias analysis of a recently published, and publicly available chest X-ray foundation model, we found reasons for concern as the model seems to encode protected characteristics including biological sex and racial identity, which may lead to disparate performance across subgroups in downstream applications. While research into foundation models for healthcare applications is in an early stage, we believe it is important to make the community aware of these risks to avoid harm.
Algorithmic encoding of protected characteristics and its implications on disparities across subgroups
Oct 27, 2021



Abstract:It has been rightfully emphasized that the use of AI for clinical decision making could amplify health disparities. A machine learning model may pick up undesirable correlations, for example, between a patient's racial identity and clinical outcome. Such correlations are often present in (historical) data used for model development. There has been an increase in studies reporting biases in disease detection models across patient subgroups. Besides the scarcity of data from underserved populations, very little is known about how these biases are encoded and how one may reduce or even remove disparate performance. There is some speculation whether algorithms may recognize patient characteristics such as biological sex or racial identity, and then directly or indirectly use this information when making predictions. But it remains unclear how we can establish whether such information is actually used. This article aims to shed some light on these issues by exploring new methodology allowing intuitive inspections of the inner working of machine learning models for image-based detection of disease. We also evaluate an effective yet debatable technique for addressing disparities leveraging the automatic prediction of patient characteristics, resulting in models with comparable true and false positive rates across subgroups. Our findings may stimulate the discussion about safe and ethical use of AI.
Transductive image segmentation: Self-training and effect of uncertainty estimation
Aug 02, 2021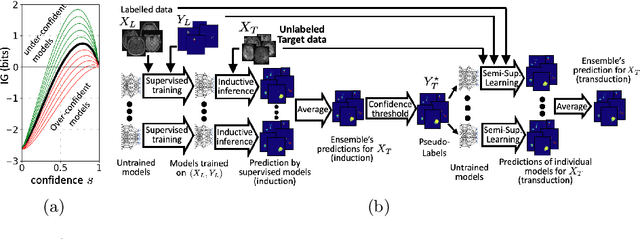

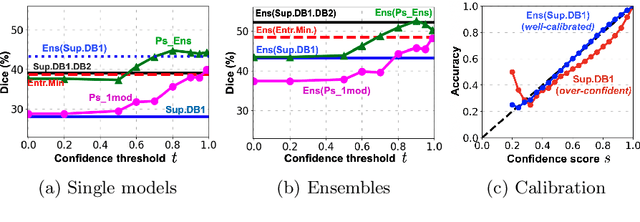
Abstract:Semi-supervised learning (SSL) uses unlabeled data during training to learn better models. Previous studies on SSL for medical image segmentation focused mostly on improving model generalization to unseen data. In some applications, however, our primary interest is not generalization but to obtain optimal predictions on a specific unlabeled database that is fully available during model development. Examples include population studies for extracting imaging phenotypes. This work investigates an often overlooked aspect of SSL, transduction. It focuses on the quality of predictions made on the unlabeled data of interest when they are included for optimization during training, rather than improving generalization. We focus on the self-training framework and explore its potential for transduction. We analyze it through the lens of Information Gain and reveal that learning benefits from the use of calibrated or under-confident models. Our extensive experiments on a large MRI database for multi-class segmentation of traumatic brain lesions shows promising results when comparing transductive with inductive predictions. We believe this study will inspire further research on transductive learning, a well-suited paradigm for medical image analysis.
Multiple Instance Learning with Auxiliary Task Weighting for Multiple Myeloma Classification
Jul 16, 2021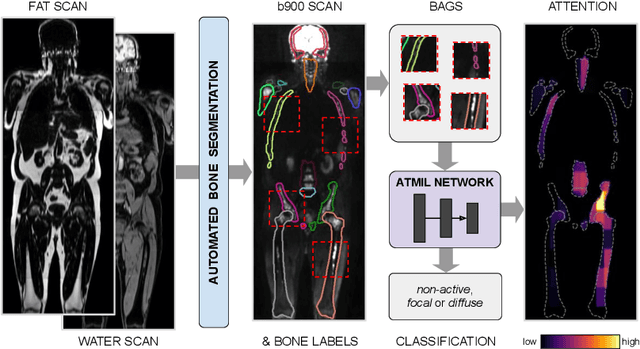
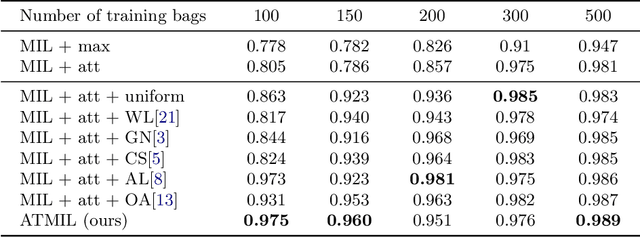
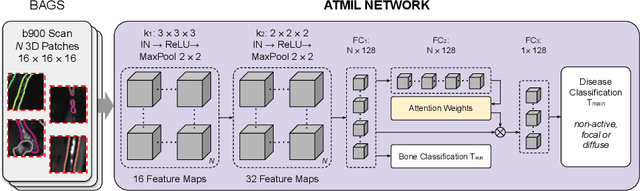
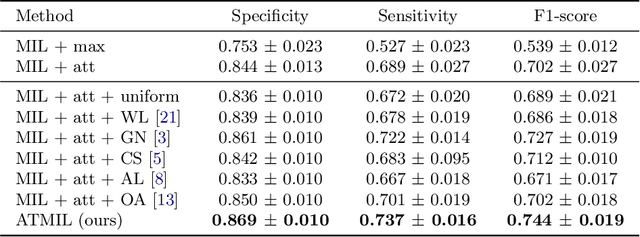
Abstract:Whole body magnetic resonance imaging (WB-MRI) is the recommended modality for diagnosis of multiple myeloma (MM). WB-MRI is used to detect sites of disease across the entire skeletal system, but it requires significant expertise and is time-consuming to report due to the great number of images. To aid radiological reading, we propose an auxiliary task-based multiple instance learning approach (ATMIL) for MM classification with the ability to localize sites of disease. This approach is appealing as it only requires patient-level annotations where an attention mechanism is used to identify local regions with active disease. We borrow ideas from multi-task learning and define an auxiliary task with adaptive reweighting to support and improve learning efficiency in the presence of data scarcity. We validate our approach on both synthetic and real multi-center clinical data. We show that the MIL attention module provides a mechanism to localize bone regions while the adaptive reweighting of the auxiliary task considerably improves the performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge