David K. Menon
The leap to ordinal: functional prognosis after traumatic brain injury using artificial intelligence
Feb 10, 2022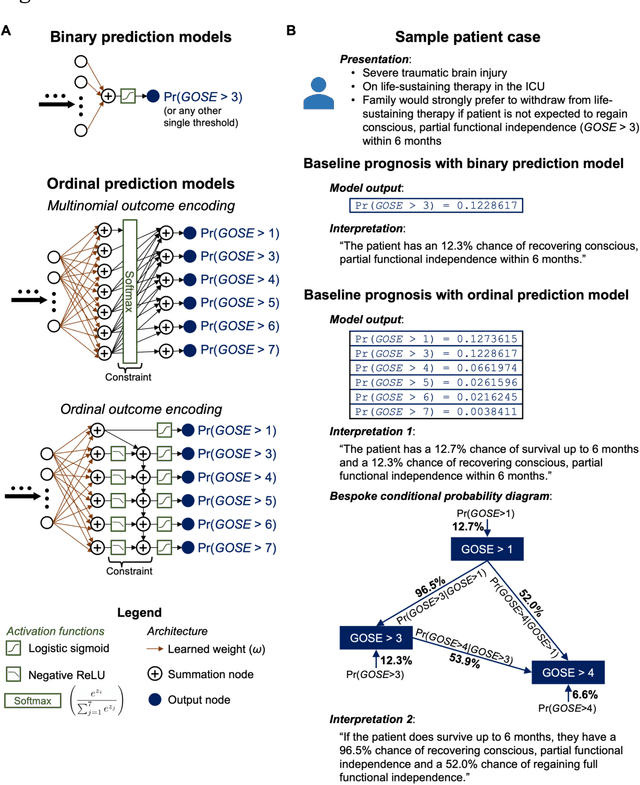
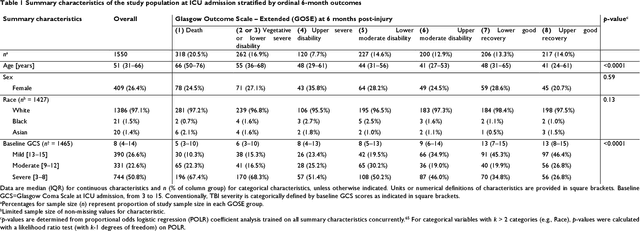
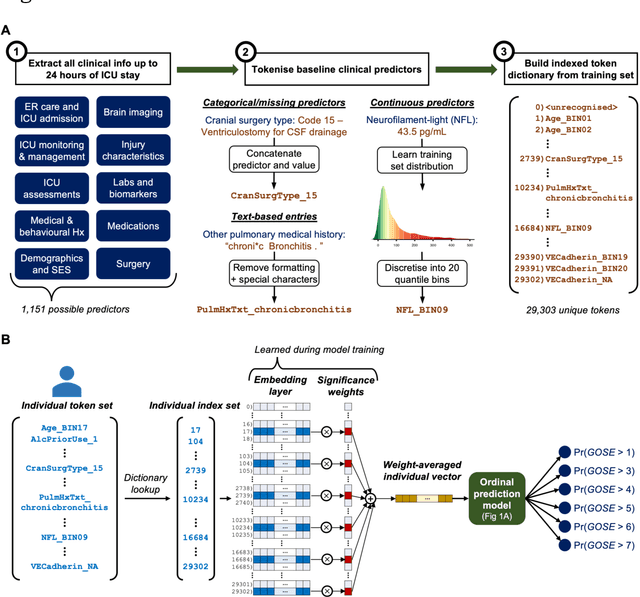
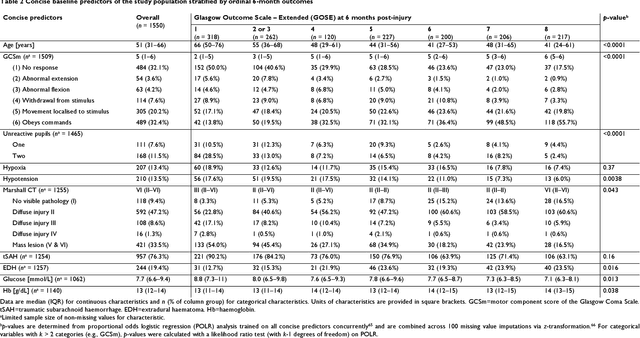
Abstract:When a patient is admitted to the intensive care unit (ICU) after a traumatic brain injury (TBI), an early prognosis is essential for baseline risk adjustment and shared decision making. TBI outcomes are commonly categorised by the Glasgow Outcome Scale-Extended (GOSE) into 8, ordered levels of functional recovery at 6 months after injury. Existing ICU prognostic models predict binary outcomes at a certain threshold of GOSE (e.g., prediction of survival [GOSE>1] or functional independence [GOSE>4]). We aimed to develop ordinal prediction models that concurrently predict probabilities of each GOSE score. From a prospective cohort (n=1,550, 65 centres) in the ICU stratum of the Collaborative European NeuroTrauma Effectiveness Research in TBI (CENTER-TBI) patient dataset, we extracted all clinical information within 24 hours of ICU admission (1,151 predictors) and 6-month GOSE scores. We analysed the effect of 2 design elements on ordinal model performance: (1) the baseline predictor set, ranging from a concise set of 10 validated predictors to a token-embedded representation of all possible predictors, and (2) the modelling strategy, from ordinal logistic regression to multinomial deep learning. With repeated k-fold cross-validation, we found that expanding the baseline predictor set significantly improved ordinal prediction performance while increasing analytical complexity did not. Half of these gains could be achieved with the addition of 8 high-impact predictors (2 demographic variables, 4 protein biomarkers, and 2 severity assessments) to the concise set. At best, ordinal models achieved 0.76 (95% CI: 0.74-0.77) ordinal discrimination ability (ordinal c-index) and 57% (95% CI: 54%-60%) explanation of ordinal variation in 6-month GOSE (Somers' D). Our results motivate the search for informative predictors for higher GOSE and the development of ordinal dynamic prediction models.
Transductive image segmentation: Self-training and effect of uncertainty estimation
Aug 02, 2021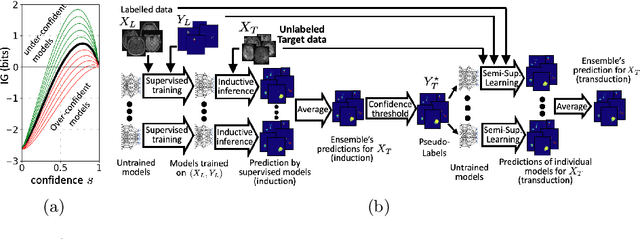

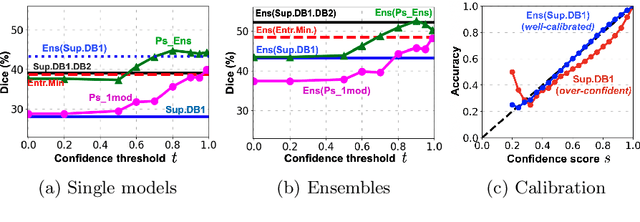
Abstract:Semi-supervised learning (SSL) uses unlabeled data during training to learn better models. Previous studies on SSL for medical image segmentation focused mostly on improving model generalization to unseen data. In some applications, however, our primary interest is not generalization but to obtain optimal predictions on a specific unlabeled database that is fully available during model development. Examples include population studies for extracting imaging phenotypes. This work investigates an often overlooked aspect of SSL, transduction. It focuses on the quality of predictions made on the unlabeled data of interest when they are included for optimization during training, rather than improving generalization. We focus on the self-training framework and explore its potential for transduction. We analyze it through the lens of Information Gain and reveal that learning benefits from the use of calibrated or under-confident models. Our extensive experiments on a large MRI database for multi-class segmentation of traumatic brain lesions shows promising results when comparing transductive with inductive predictions. We believe this study will inspire further research on transductive learning, a well-suited paradigm for medical image analysis.
Efficient Multi-Scale 3D CNN with Fully Connected CRF for Accurate Brain Lesion Segmentation
Jan 08, 2017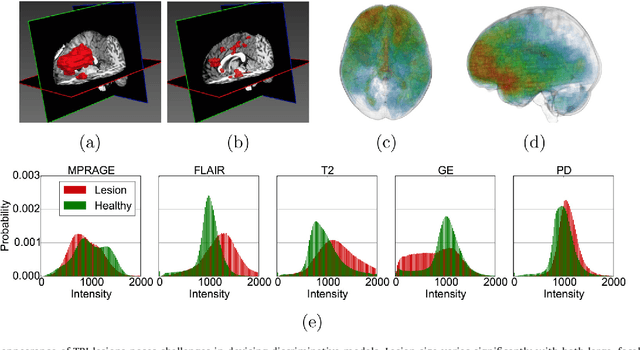
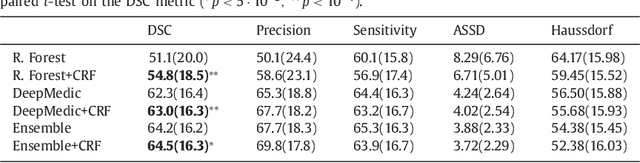
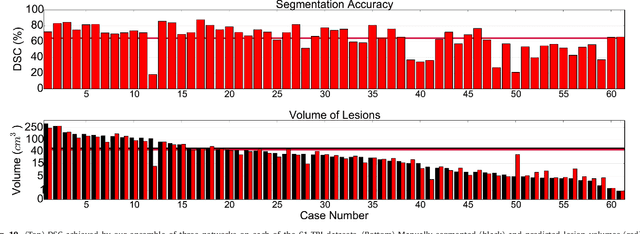
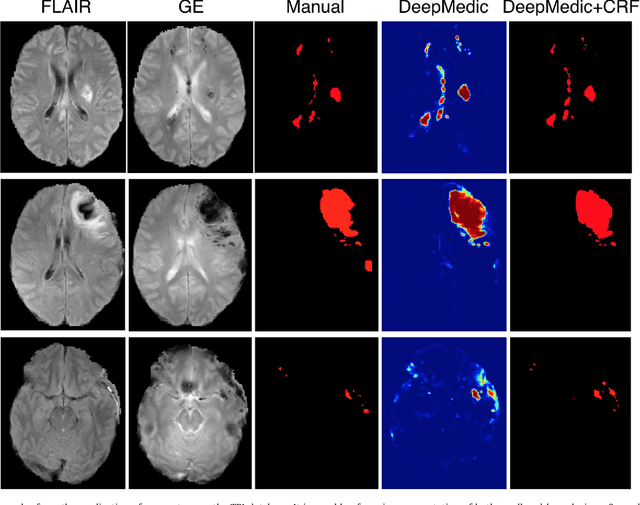
Abstract:We propose a dual pathway, 11-layers deep, three-dimensional Convolutional Neural Network for the challenging task of brain lesion segmentation. The devised architecture is the result of an in-depth analysis of the limitations of current networks proposed for similar applications. To overcome the computational burden of processing 3D medical scans, we have devised an efficient and effective dense training scheme which joins the processing of adjacent image patches into one pass through the network while automatically adapting to the inherent class imbalance present in the data. Further, we analyze the development of deeper, thus more discriminative 3D CNNs. In order to incorporate both local and larger contextual information, we employ a dual pathway architecture that processes the input images at multiple scales simultaneously. For post-processing of the network's soft segmentation, we use a 3D fully connected Conditional Random Field which effectively removes false positives. Our pipeline is extensively evaluated on three challenging tasks of lesion segmentation in multi-channel MRI patient data with traumatic brain injuries, brain tumors, and ischemic stroke. We improve on the state-of-the-art for all three applications, with top ranking performance on the public benchmarks BRATS 2015 and ISLES 2015. Our method is computationally efficient, which allows its adoption in a variety of research and clinical settings. The source code of our implementation is made publicly available.
Unsupervised domain adaptation in brain lesion segmentation with adversarial networks
Dec 28, 2016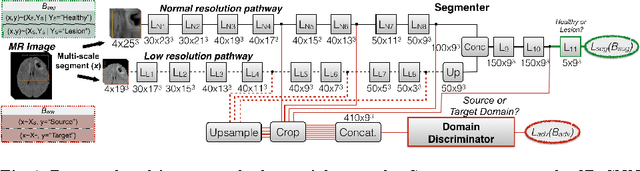
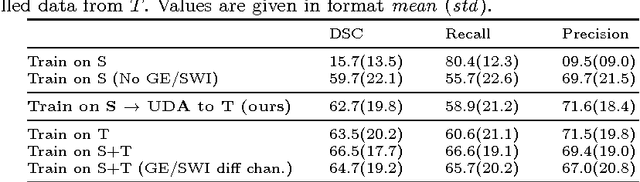
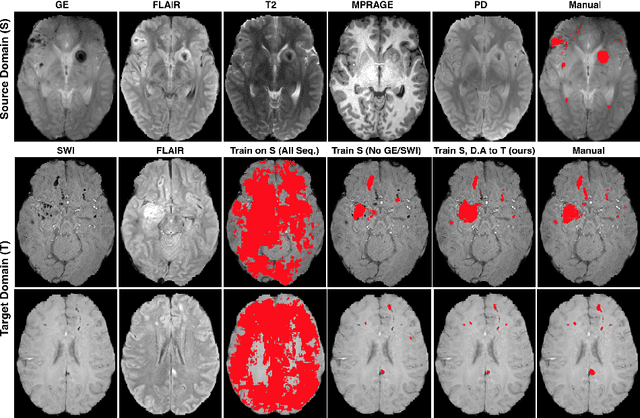
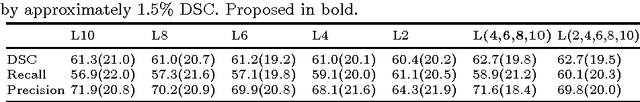
Abstract:Significant advances have been made towards building accurate automatic segmentation systems for a variety of biomedical applications using machine learning. However, the performance of these systems often degrades when they are applied on new data that differ from the training data, for example, due to variations in imaging protocols. Manually annotating new data for each test domain is not a feasible solution. In this work we investigate unsupervised domain adaptation using adversarial neural networks to train a segmentation method which is more invariant to differences in the input data, and which does not require any annotations on the test domain. Specifically, we learn domain-invariant features by learning to counter an adversarial network, which attempts to classify the domain of the input data by observing the activations of the segmentation network. Furthermore, we propose a multi-connected domain discriminator for improved adversarial training. Our system is evaluated using two MR databases of subjects with traumatic brain injuries, acquired using different scanners and imaging protocols. Using our unsupervised approach, we obtain segmentation accuracies which are close to the upper bound of supervised domain adaptation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge