Christian Baumgartner
CUTE-MRI: Conformalized Uncertainty-based framework for Time-adaptivE MRI
Aug 20, 2025Abstract:Magnetic Resonance Imaging (MRI) offers unparalleled soft-tissue contrast but is fundamentally limited by long acquisition times. While deep learning-based accelerated MRI can dramatically shorten scan times, the reconstruction from undersampled data introduces ambiguity resulting from an ill-posed problem with infinitely many possible solutions that propagates to downstream clinical tasks. This uncertainty is usually ignored during the acquisition process as acceleration factors are often fixed a priori, resulting in scans that are either unnecessarily long or of insufficient quality for a given clinical endpoint. This work introduces a dynamic, uncertainty-aware acquisition framework that adjusts scan time on a per-subject basis. Our method leverages a probabilistic reconstruction model to estimate image uncertainty, which is then propagated through a full analysis pipeline to a quantitative metric of interest (e.g., patellar cartilage volume or cardiac ejection fraction). We use conformal prediction to transform this uncertainty into a rigorous, calibrated confidence interval for the metric. During acquisition, the system iteratively samples k-space, updates the reconstruction, and evaluates the confidence interval. The scan terminates automatically once the uncertainty meets a user-predefined precision target. We validate our framework on both knee and cardiac MRI datasets. Our results demonstrate that this adaptive approach reduces scan times compared to fixed protocols while providing formal statistical guarantees on the precision of the final image. This framework moves beyond fixed acceleration factors, enabling patient-specific acquisitions that balance scan efficiency with diagnostic confidence, a critical step towards personalized and resource-efficient MRI.
A Human-Centered Machine-Learning Approach for Muscle-Tendon Junction Tracking in Ultrasound Images
Feb 10, 2022



Abstract:Biomechanical and clinical gait research observes muscles and tendons in limbs to study their functions and behaviour. Therefore, movements of distinct anatomical landmarks, such as muscle-tendon junctions, are frequently measured. We propose a reliable and time efficient machine-learning approach to track these junctions in ultrasound videos and support clinical biomechanists in gait analysis. In order to facilitate this process, a method based on deep-learning was introduced. We gathered an extensive dataset, covering 3 functional movements, 2 muscles, collected on 123 healthy and 38 impaired subjects with 3 different ultrasound systems, and providing a total of 66864 annotated ultrasound images in our network training. Furthermore, we used data collected across independent laboratories and curated by researchers with varying levels of experience. For the evaluation of our method a diverse test-set was selected that is independently verified by four specialists. We show that our model achieves similar performance scores to the four human specialists in identifying the muscle-tendon junction position. Our method provides time-efficient tracking of muscle-tendon junctions, with prediction times of up to 0.078 seconds per frame (approx. 100 times faster than manual labeling). All our codes, trained models and test-set were made publicly available and our model is provided as a free-to-use online service on https://deepmtj.org/.
Automatic Tracking of the Muscle Tendon Junction in Healthy and Impaired Subjects using Deep Learning
May 05, 2020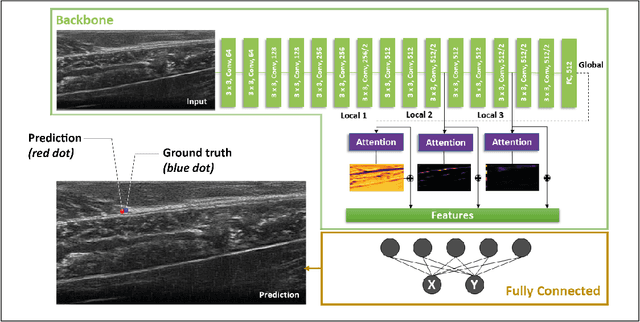



Abstract:Recording muscle tendon junction displacements during movement, allows separate investigation of the muscle and tendon behaviour, respectively. In order to provide a fully-automatic tracking method, we employ a novel deep learning approach to detect the position of the muscle tendon junction in ultrasound images. We utilize the attention mechanism to enable the network to focus on relevant regions and to obtain a better interpretation of the results. Our data set consists of a large cohort of 79 healthy subjects and 28 subjects with movement limitations performing passive full range of motion and maximum contraction movements. Our trained network shows robust detection of the muscle tendon junction on a diverse data set of varying quality with a mean absolute error of 2.55$\pm$1 mm. We show that our approach can be applied for various subjects and can be operated in real-time. The complete software package is available for open-source use via: https://github.com/luuleitner/deepMTJ
Semi-Supervised and Task-Driven Data Augmentation
Feb 28, 2019



Abstract:Supervised deep learning methods for segmentation require large amounts of labelled training data, without which they are prone to overfitting, not generalizing well to unseen images. In practice, obtaining a large number of annotations from clinical experts is expensive and time-consuming. One way to address scarcity of annotated examples is data augmentation using random spatial and intensity transformations. Recently, it has been proposed to use generative models to synthesize realistic training examples, complementing the random augmentation. So far, these methods have yielded limited gains over the random augmentation. However, there is potential to improve the approach by (i) explicitly modeling deformation fields (non-affine spatial transformation) and intensity transformations and (ii) leveraging unlabelled data during the generative process. With this motivation, we propose a novel task-driven data augmentation method where to synthesize new training examples, a generative network explicitly models and applies deformation fields and additive intensity masks on existing labelled data, modeling shape and intensity variations, respectively. Crucially, the generative model is optimized to be conducive to the task, in this case segmentation, and constrained to match the distribution of images observed from labelled and unlabelled samples. Furthermore, explicit modeling of deformation fields allow synthesizing segmentation masks and images in exact correspondence by simply applying the generated transformation to an input image and the corresponding annotation. Our experiments on cardiac magnetic resonance images (MRI) showed that, for the task of segmentation in small training data scenarios, the proposed method substantially outperforms conventional augmentation techniques.
A Lifelong Learning Approach to Brain MR Segmentation Across Scanners and Protocols
May 25, 2018



Abstract:Convolutional neural networks (CNNs) have shown promising results on several segmentation tasks in magnetic resonance (MR) images. However, the accuracy of CNNs may degrade severely when segmenting images acquired with different scanners and/or protocols as compared to the training data, thus limiting their practical utility. We address this shortcoming in a lifelong multi-domain learning setting by treating images acquired with different scanners or protocols as samples from different, but related domains. Our solution is a single CNN with shared convolutional filters and domain-specific batch normalization layers, which can be tuned to new domains with only a few ($\approx$ 4) labelled images. Importantly, this is achieved while retaining performance on the older domains whose training data may no longer be available. We evaluate the method for brain structure segmentation in MR images. Results demonstrate that the proposed method largely closes the gap to the benchmark, which is training a dedicated CNN for each scanner.
Unsupervised domain adaptation in brain lesion segmentation with adversarial networks
Dec 28, 2016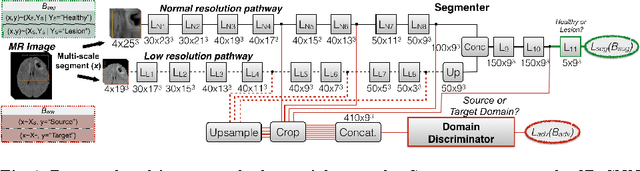
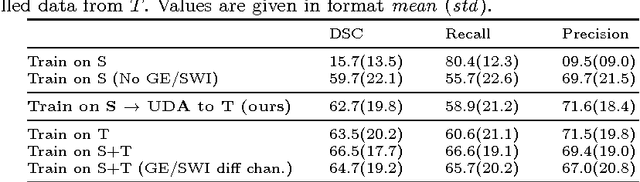
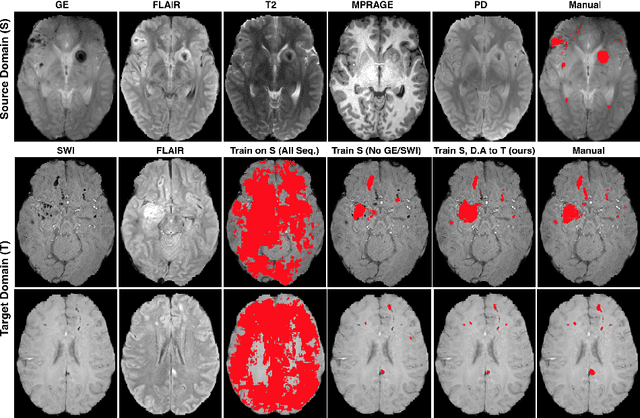
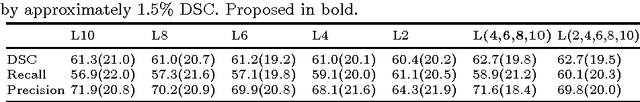
Abstract:Significant advances have been made towards building accurate automatic segmentation systems for a variety of biomedical applications using machine learning. However, the performance of these systems often degrades when they are applied on new data that differ from the training data, for example, due to variations in imaging protocols. Manually annotating new data for each test domain is not a feasible solution. In this work we investigate unsupervised domain adaptation using adversarial neural networks to train a segmentation method which is more invariant to differences in the input data, and which does not require any annotations on the test domain. Specifically, we learn domain-invariant features by learning to counter an adversarial network, which attempts to classify the domain of the input data by observing the activations of the segmentation network. Furthermore, we propose a multi-connected domain discriminator for improved adversarial training. Our system is evaluated using two MR databases of subjects with traumatic brain injuries, acquired using different scanners and imaging protocols. Using our unsupervised approach, we obtain segmentation accuracies which are close to the upper bound of supervised domain adaptation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge