Antonio Criminisi
Hairmony: Fairness-aware hairstyle classification
Oct 15, 2024



Abstract:We present a method for prediction of a person's hairstyle from a single image. Despite growing use cases in user digitization and enrollment for virtual experiences, available methods are limited, particularly in the range of hairstyles they can capture. Human hair is extremely diverse and lacks any universally accepted description or categorization, making this a challenging task. Most current methods rely on parametric models of hair at a strand level. These approaches, while very promising, are not yet able to represent short, frizzy, coily hair and gathered hairstyles. We instead choose a classification approach which can represent the diversity of hairstyles required for a truly robust and inclusive system. Previous classification approaches have been restricted by poorly labeled data that lacks diversity, imposing constraints on the usefulness of any resulting enrollment system. We use only synthetic data to train our models. This allows for explicit control of diversity of hairstyle attributes, hair colors, facial appearance, poses, environments and other parameters. It also produces noise-free ground-truth labels. We introduce a novel hairstyle taxonomy developed in collaboration with a diverse group of domain experts which we use to balance our training data, supervise our model, and directly measure fairness. We annotate our synthetic training data and a real evaluation dataset using this taxonomy and release both to enable comparison of future hairstyle prediction approaches. We employ an architecture based on a pre-trained feature extraction network in order to improve generalization of our method to real data and predict taxonomy attributes as an auxiliary task to improve accuracy. Results show our method to be significantly more robust for challenging hairstyles than recent parametric approaches.
Sequential Ensembling for Semantic Segmentation
Oct 08, 2022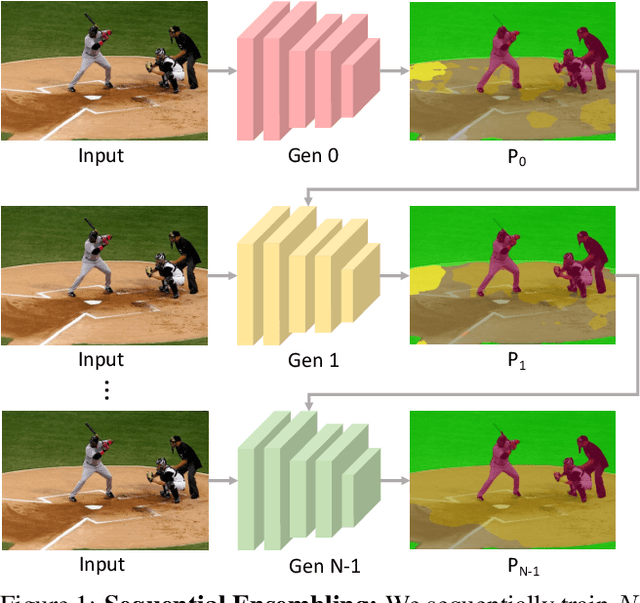
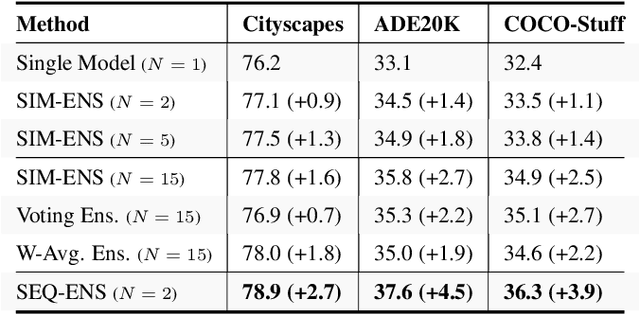
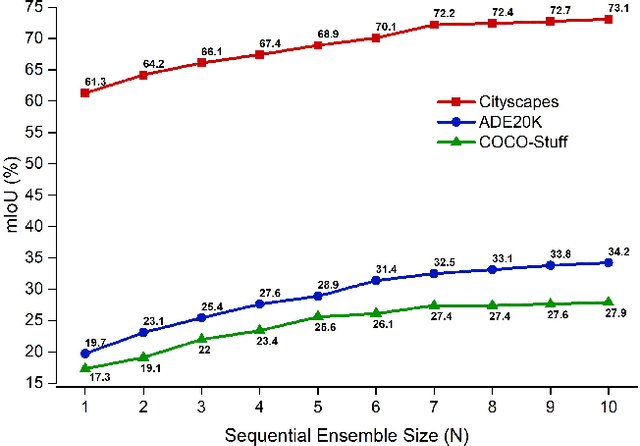
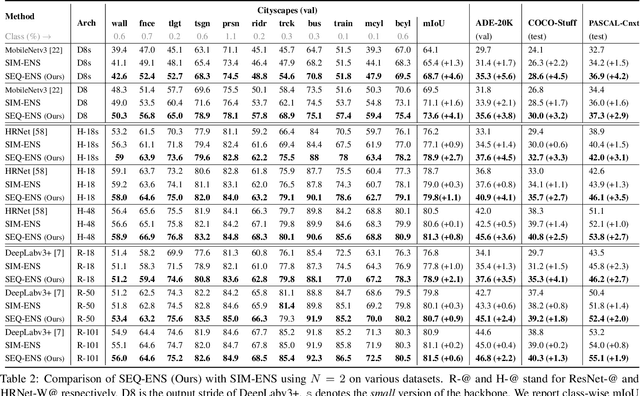
Abstract:Ensemble approaches for deep-learning-based semantic segmentation remain insufficiently explored despite the proliferation of competitive benchmarks and downstream applications. In this work, we explore and benchmark the popular ensembling approach of combining predictions of multiple, independently-trained, state-of-the-art models at test time on popular datasets. Furthermore, we propose a novel method inspired by boosting to sequentially ensemble networks that significantly outperforms the naive ensemble baseline. Our approach trains a cascade of models conditioned on class probabilities predicted by the previous model as an additional input. A key benefit of this approach is that it allows for dynamic computation offloading, which helps deploy models on mobile devices. Our proposed novel ADaptive modulatiON (ADON) block allows spatial feature modulation at various layers using previous-stage probabilities. Our approach does not require sophisticated sample selection strategies during training and works with multiple neural architectures. We significantly improve over the naive ensemble baseline on challenging datasets such as Cityscapes, ADE-20K, COCO-Stuff, and PASCAL-Context and set a new state-of-the-art.
Uncertainty Quantification in Deep Learning for Safer Neuroimage Enhancement
Jul 31, 2019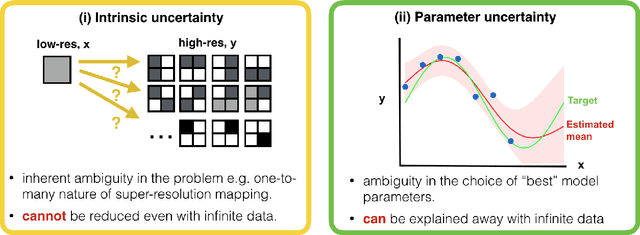
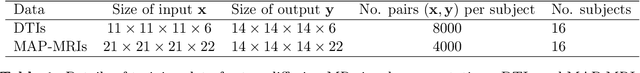
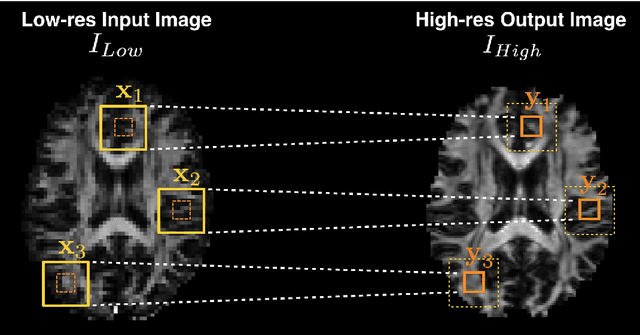
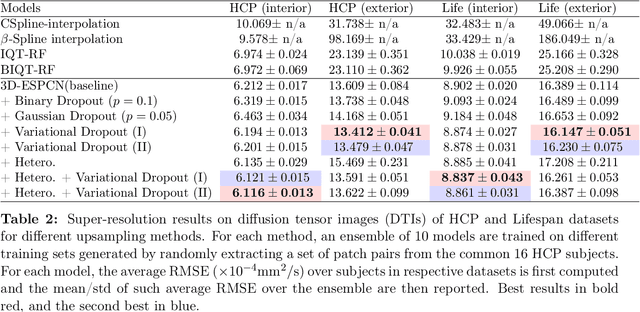
Abstract:Deep learning (DL) has shown great potential in medical image enhancement problems, such as super-resolution or image synthesis. However, to date, little consideration has been given to uncertainty quantification over the output image. Here we introduce methods to characterise different components of uncertainty in such problems and demonstrate the ideas using diffusion MRI super-resolution. Specifically, we propose to account for $intrinsic$ uncertainty through a heteroscedastic noise model and for $parameter$ uncertainty through approximate Bayesian inference, and integrate the two to quantify $predictive$ uncertainty over the output image. Moreover, we introduce a method to propagate the predictive uncertainty on a multi-channelled image to derived scalar parameters, and separately quantify the effects of intrinsic and parameter uncertainty therein. The methods are evaluated for super-resolution of two different signal representations of diffusion MR images---DTIs and Mean Apparent Propagator MRI---and their derived quantities such as MD and FA, on multiple datasets of both healthy and pathological human brains. Results highlight three key benefits of uncertainty modelling for improving the safety of DL-based image enhancement systems. Firstly, incorporating uncertainty improves the predictive performance even when test data departs from training data. Secondly, the predictive uncertainty highly correlates with errors, and is therefore capable of detecting predictive "failures". Results demonstrate that such an uncertainty measure enables subject-specific and voxel-wise risk assessment of the output images. Thirdly, we show that the method for decomposing predictive uncertainty into its independent sources provides high-level "explanations" for the performance by quantifying how much uncertainty arises from the inherent difficulty of the task or the limited training examples.
Deep Learning with Mixed Supervision for Brain Tumor Segmentation
Dec 10, 2018



Abstract:Most of the current state-of-the-art methods for tumor segmentation are based on machine learning models trained on manually segmented images. This type of training data is particularly costly, as manual delineation of tumors is not only time-consuming but also requires medical expertise. On the other hand, images with a provided global label (indicating presence or absence of a tumor) are less informative but can be obtained at a substantially lower cost. In this paper, we propose to use both types of training data (fully-annotated and weakly-annotated) to train a deep learning model for segmentation. The idea of our approach is to extend segmentation networks with an additional branch performing image-level classification. The model is jointly trained for segmentation and classification tasks in order to exploit information contained in weakly-annotated images while preventing the network to learn features which are irrelevant for the segmentation task. We evaluate our method on the challenging task of brain tumor segmentation in Magnetic Resonance images from BRATS 2018 challenge. We show that the proposed approach provides a significant improvement of segmentation performance compared to the standard supervised learning. The observed improvement is proportional to the ratio between weakly-annotated and fully-annotated images available for training.
Adaptive Neural Trees
Oct 07, 2018
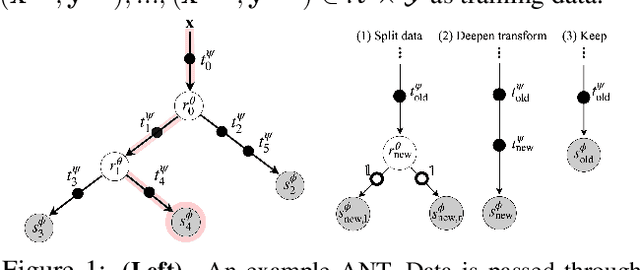


Abstract:Deep neural networks and decision trees operate on largely separate paradigms; typically, the former performs representation learning with pre-specified architectures, while the latter is characterised by learning hierarchies over pre-specified features with data-driven architectures. We unite the two via adaptive neural trees (ANTs), a model that incorporates representation learning into edges, routing functions and leaf nodes of a decision tree, along with a backpropagation-based training algorithm that adaptively grows the architecture from primitive modules (e.g., convolutional layers). ANTs allow increased interpretability via hierarchical clustering, e.g., learning meaningful class associations, such as separating natural vs. man-made objects. We demonstrate this whilst achieving over 99% and 90% accuracy on the MNIST and CIFAR-10 datasets. Furthermore, ANT optimisation naturally adapts the architecture to the size and complexity of the training data.
Semi-Supervised Learning via Compact Latent Space Clustering
Jul 29, 2018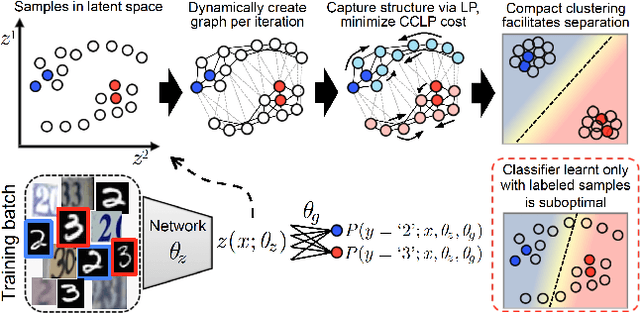
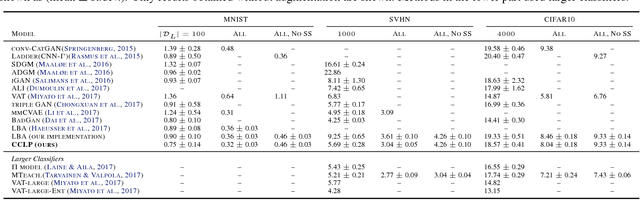
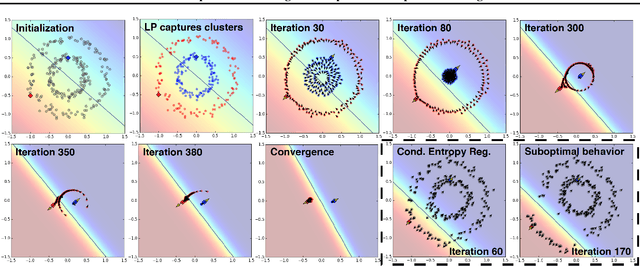
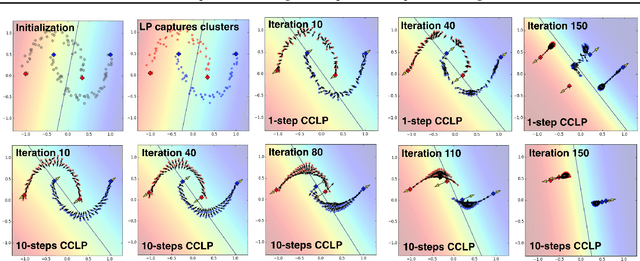
Abstract:We present a novel cost function for semi-supervised learning of neural networks that encourages compact clustering of the latent space to facilitate separation. The key idea is to dynamically create a graph over embeddings of labeled and unlabeled samples of a training batch to capture underlying structure in feature space, and use label propagation to estimate its high and low density regions. We then devise a cost function based on Markov chains on the graph that regularizes the latent space to form a single compact cluster per class, while avoiding to disturb existing clusters during optimization. We evaluate our approach on three benchmarks and compare to state-of-the art with promising results. Our approach combines the benefits of graph-based regularization with efficient, inductive inference, does not require modifications to a network architecture, and can thus be easily applied to existing networks to enable an effective use of unlabeled data.
3D Convolutional Neural Networks for Tumor Segmentation using Long-range 2D Context
Jul 23, 2018



Abstract:We present an efficient deep learning approach for the challenging task of tumor segmentation in multisequence MR images. In recent years, Convolutional Neural Networks (CNN) have achieved state-of-the-art performances in a large variety of recognition tasks in medical imaging. Because of the considerable computational cost of CNNs, large volumes such as MRI are typically processed by subvolumes, for instance slices (axial, coronal, sagittal) or small 3D patches. In this paper we introduce a CNN-based model which efficiently combines the advantages of the short-range 3D context and the long-range 2D context. To overcome the limitations of specific choices of neural network architectures, we also propose to merge outputs of several cascaded 2D-3D models by a voxelwise voting strategy. Furthermore, we propose a network architecture in which the different MR sequences are processed by separate subnetworks in order to be more robust to the problem of missing MR sequences. Finally, a simple and efficient algorithm for training large CNN models is introduced. We evaluate our method on the public benchmark of the BRATS 2017 challenge on the task of multiclass segmentation of malignant brain tumors. Our method achieves good performances and produces accurate segmentations with median Dice scores of 0.918 (whole tumor), 0.883 (tumor core) and 0.854 (enhancing core). Our approach can be naturally applied to various tasks involving segmentation of lesions or organs.
Autofocus Layer for Semantic Segmentation
Jun 11, 2018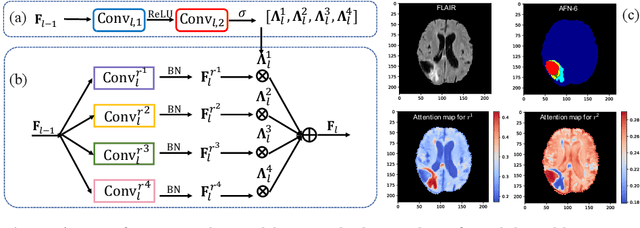
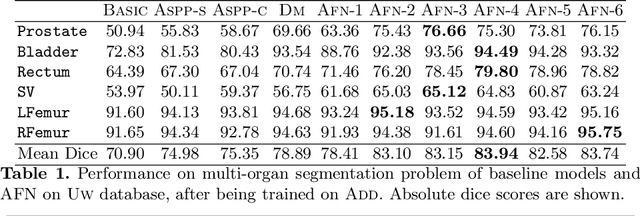
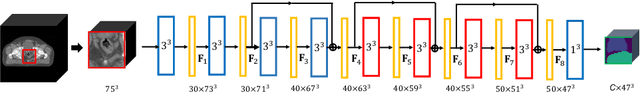
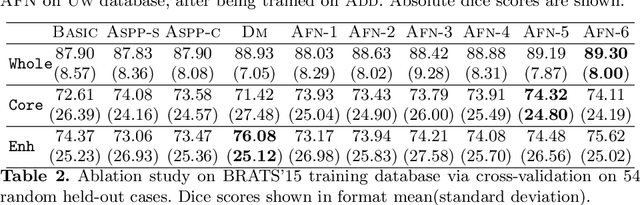
Abstract:We propose the autofocus convolutional layer for semantic segmentation with the objective of enhancing the capabilities of neural networks for multi-scale processing. Autofocus layers adaptively change the size of the effective receptive field based on the processed context to generate more powerful features. This is achieved by parallelising multiple convolutional layers with different dilation rates, combined by an attention mechanism that learns to focus on the optimal scales driven by context. By sharing the weights of the parallel convolutions we make the network scale-invariant, with only a modest increase in the number of parameters. The proposed autofocus layer can be easily integrated into existing networks to improve a model's representational power. We evaluate our models on the challenging tasks of multi-organ segmentation in pelvic CT and brain tumor segmentation in MRI and achieve very promising performance.
Measuring Neural Net Robustness with Constraints
Jun 16, 2017
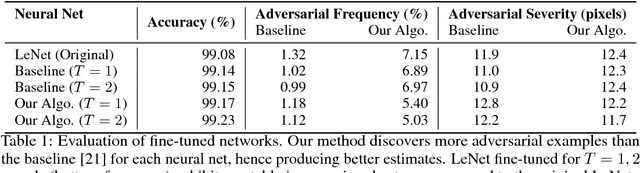

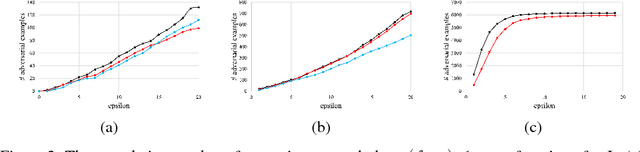
Abstract:Despite having high accuracy, neural nets have been shown to be susceptible to adversarial examples, where a small perturbation to an input can cause it to become mislabeled. We propose metrics for measuring the robustness of a neural net and devise a novel algorithm for approximating these metrics based on an encoding of robustness as a linear program. We show how our metrics can be used to evaluate the robustness of deep neural nets with experiments on the MNIST and CIFAR-10 datasets. Our algorithm generates more informative estimates of robustness metrics compared to estimates based on existing algorithms. Furthermore, we show how existing approaches to improving robustness "overfit" to adversarial examples generated using a specific algorithm. Finally, we show that our techniques can be used to additionally improve neural net robustness both according to the metrics that we propose, but also according to previously proposed metrics.
Bayesian Image Quality Transfer with CNNs: Exploring Uncertainty in dMRI Super-Resolution
May 30, 2017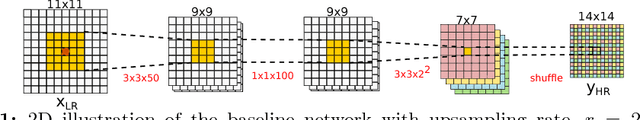
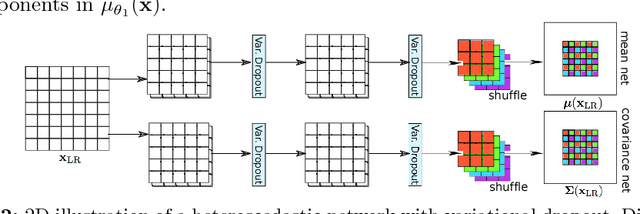
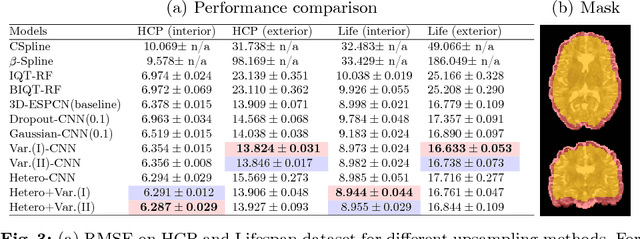
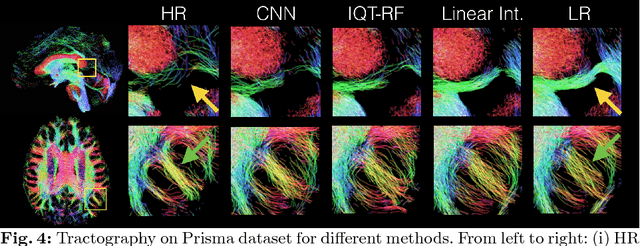
Abstract:In this work, we investigate the value of uncertainty modeling in 3D super-resolution with convolutional neural networks (CNNs). Deep learning has shown success in a plethora of medical image transformation problems, such as super-resolution (SR) and image synthesis. However, the highly ill-posed nature of such problems results in inevitable ambiguity in the learning of networks. We propose to account for intrinsic uncertainty through a per-patch heteroscedastic noise model and for parameter uncertainty through approximate Bayesian inference in the form of variational dropout. We show that the combined benefits of both lead to the state-of-the-art performance SR of diffusion MR brain images in terms of errors compared to ground truth. We further show that the reduced error scores produce tangible benefits in downstream tractography. In addition, the probabilistic nature of the methods naturally confers a mechanism to quantify uncertainty over the super-resolved output. We demonstrate through experiments on both healthy and pathological brains the potential utility of such an uncertainty measure in the risk assessment of the super-resolved images for subsequent clinical use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge