Jay Nanavati
Agentic AI for Scientific Discovery: A Survey of Progress, Challenges, and Future Directions
Mar 12, 2025Abstract:The integration of Agentic AI into scientific discovery marks a new frontier in research automation. These AI systems, capable of reasoning, planning, and autonomous decision-making, are transforming how scientists perform literature review, generate hypotheses, conduct experiments, and analyze results. This survey provides a comprehensive overview of Agentic AI for scientific discovery, categorizing existing systems and tools, and highlighting recent progress across fields such as chemistry, biology, and materials science. We discuss key evaluation metrics, implementation frameworks, and commonly used datasets to offer a detailed understanding of the current state of the field. Finally, we address critical challenges, such as literature review automation, system reliability, and ethical concerns, while outlining future research directions that emphasize human-AI collaboration and enhanced system calibration.
OntologyRAG: Better and Faster Biomedical Code Mapping with Retrieval-Augmented Generation (RAG) Leveraging Ontology Knowledge Graphs and Large Language Models
Feb 26, 2025



Abstract:Biomedical ontologies, which comprehensively define concepts and relations for biomedical entities, are crucial for structuring and formalizing domain-specific information representations. Biomedical code mapping identifies similarity or equivalence between concepts from different ontologies. Obtaining high-quality mapping usually relies on automatic generation of unrefined mapping with ontology domain fine-tuned language models (LMs), followed by manual selections or corrections by coding experts who have extensive domain expertise and familiarity with ontology schemas. The LMs usually provide unrefined code mapping suggestions as a list of candidates without reasoning or supporting evidence, hence coding experts still need to verify each suggested candidate against ontology sources to pick the best matches. This is also a recurring task as ontology sources are updated regularly to incorporate new research findings. Consequently, the need of regular LM retraining and manual refinement make code mapping time-consuming and labour intensive. In this work, we created OntologyRAG, an ontology-enhanced retrieval-augmented generation (RAG) method that leverages the inductive biases from ontological knowledge graphs for in-context-learning (ICL) in large language models (LLMs). Our solution grounds LLMs to knowledge graphs with unrefined mappings between ontologies and processes questions by generating an interpretable set of results that include prediction rational with mapping proximity assessment. Our solution doesn't require re-training LMs, as all ontology updates could be reflected by updating the knowledge graphs with a standard process. Evaluation results on a self-curated gold dataset show promises of using our method to enable coding experts to achieve better and faster code mapping. The code is available at https://github.com/iqvianlp/ontologyRAG.
Towards Ontology-Enhanced Representation Learning for Large Language Models
May 30, 2024Abstract:Taking advantage of the widespread use of ontologies to organise and harmonize knowledge across several distinct domains, this paper proposes a novel approach to improve an embedding-Large Language Model (embedding-LLM) of interest by infusing the knowledge formalized by a reference ontology: ontological knowledge infusion aims at boosting the ability of the considered LLM to effectively model the knowledge domain described by the infused ontology. The linguistic information (i.e. concept synonyms and descriptions) and structural information (i.e. is-a relations) formalized by the ontology are utilized to compile a comprehensive set of concept definitions, with the assistance of a powerful generative LLM (i.e. GPT-3.5-turbo). These concept definitions are then employed to fine-tune the target embedding-LLM using a contrastive learning framework. To demonstrate and evaluate the proposed approach, we utilize the biomedical disease ontology MONDO. The results show that embedding-LLMs enhanced by ontological disease knowledge exhibit an improved capability to effectively evaluate the similarity of in-domain sentences from biomedical documents mentioning diseases, without compromising their out-of-domain performance.
Autofocus Layer for Semantic Segmentation
Jun 11, 2018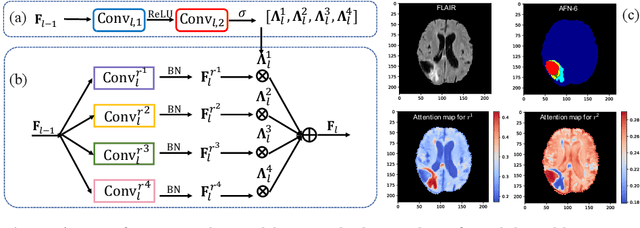
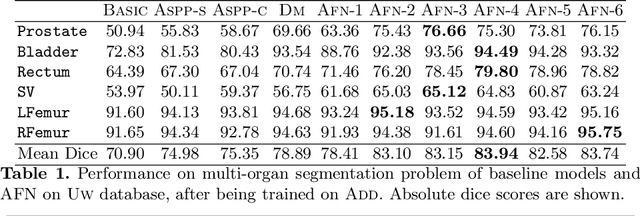
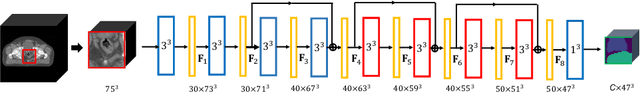
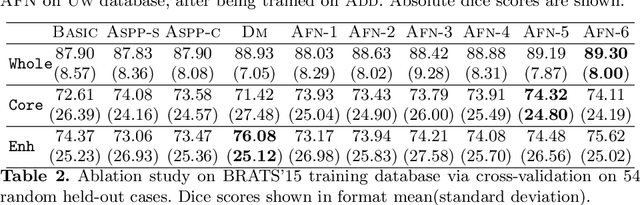
Abstract:We propose the autofocus convolutional layer for semantic segmentation with the objective of enhancing the capabilities of neural networks for multi-scale processing. Autofocus layers adaptively change the size of the effective receptive field based on the processed context to generate more powerful features. This is achieved by parallelising multiple convolutional layers with different dilation rates, combined by an attention mechanism that learns to focus on the optimal scales driven by context. By sharing the weights of the parallel convolutions we make the network scale-invariant, with only a modest increase in the number of parameters. The proposed autofocus layer can be easily integrated into existing networks to improve a model's representational power. We evaluate our models on the challenging tasks of multi-organ segmentation in pelvic CT and brain tumor segmentation in MRI and achieve very promising performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge