Nicholas Ayache
EPIONE
Spatial regularisation for improved accuracy and interpretability in keypoint-based registration
Mar 07, 2025Abstract:Unsupervised registration strategies bypass requirements in ground truth transforms or segmentations by optimising similarity metrics between fixed and moved volumes. Among these methods, a recent subclass of approaches based on unsupervised keypoint detection stand out as very promising for interpretability. Specifically, these methods train a network to predict feature maps for fixed and moving images, from which explainable centres of mass are computed to obtain point clouds, that are then aligned in closed-form. However, the features returned by the network often yield spatially diffuse patterns that are hard to interpret, thus undermining the purpose of keypoint-based registration. Here, we propose a three-fold loss to regularise the spatial distribution of the features. First, we use the KL divergence to model features as point spread functions that we interpret as probabilistic keypoints. Then, we sharpen the spatial distributions of these features to increase the precision of the detected landmarks. Finally, we introduce a new repulsive loss across keypoints to encourage spatial diversity. Overall, our loss considerably improves the interpretability of the features, which now correspond to precise and anatomically meaningful landmarks. We demonstrate our three-fold loss in foetal rigid motion tracking and brain MRI affine registration tasks, where it not only outperforms state-of-the-art unsupervised strategies, but also bridges the gap with state-of-the-art supervised methods. Our code is available at https://github.com/BenBillot/spatial_regularisation.
Generative Medical Image Anonymization Based on Latent Code Projection and Optimization
Jan 15, 2025Abstract:Medical image anonymization aims to protect patient privacy by removing identifying information, while preserving the data utility to solve downstream tasks. In this paper, we address the medical image anonymization problem with a two-stage solution: latent code projection and optimization. In the projection stage, we design a streamlined encoder to project input images into a latent space and propose a co-training scheme to enhance the projection process. In the optimization stage, we refine the latent code using two deep loss functions designed to address the trade-off between identity protection and data utility dedicated to medical images. Through a comprehensive set of qualitative and quantitative experiments, we showcase the effectiveness of our approach on the MIMIC-CXR chest X-ray dataset by generating anonymized synthetic images that can serve as training set for detecting lung pathologies. Source codes are available at https://github.com/Huiyu-Li/GMIA.
Morphologically-Aware Consensus Computation via Heuristics-based IterATive Optimization (MACCHIatO)
Sep 14, 2023Abstract:The extraction of consensus segmentations from several binary or probabilistic masks is important to solve various tasks such as the analysis of inter-rater variability or the fusion of several neural network outputs. One of the most widely used methods to obtain such a consensus segmentation is the STAPLE algorithm. In this paper, we first demonstrate that the output of that algorithm is heavily impacted by the background size of images and the choice of the prior. We then propose a new method to construct a binary or a probabilistic consensus segmentation based on the Fr\'{e}chet means of carefully chosen distances which makes it totally independent of the image background size. We provide a heuristic approach to optimize this criterion such that a voxel's class is fully determined by its voxel-wise distance to the different masks, the connected component it belongs to and the group of raters who segmented it. We compared extensively our method on several datasets with the STAPLE method and the naive segmentation averaging method, showing that it leads to binary consensus masks of intermediate size between Majority Voting and STAPLE and to different posterior probabilities than Mask Averaging and STAPLE methods. Our code is available at https://gitlab.inria.fr/dhamzaou/jaccardmap .
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:013
Real-Time Dynamic Data Driven Deformable Registration for Image-Guided Neurosurgery: Computational Aspects
Sep 06, 2023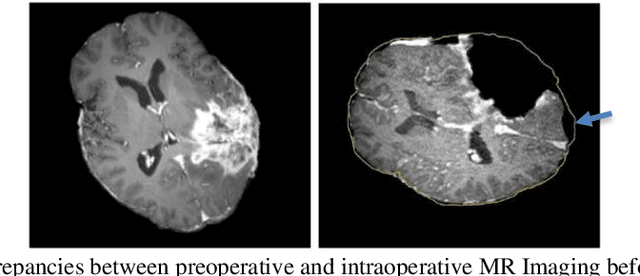
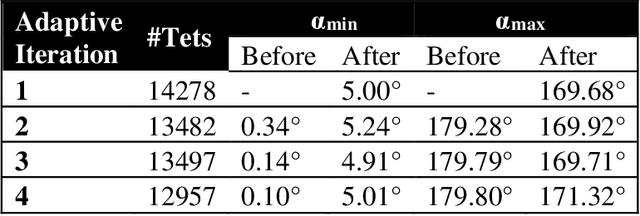
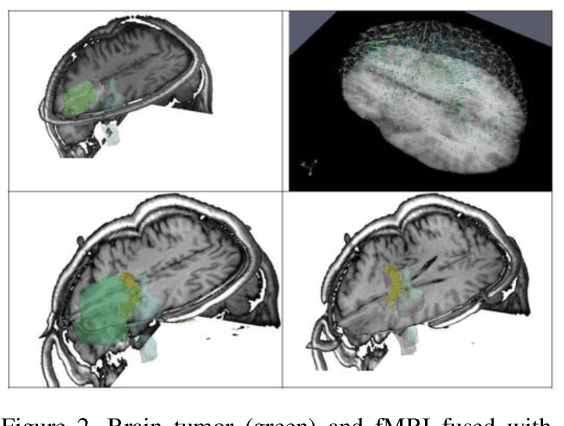
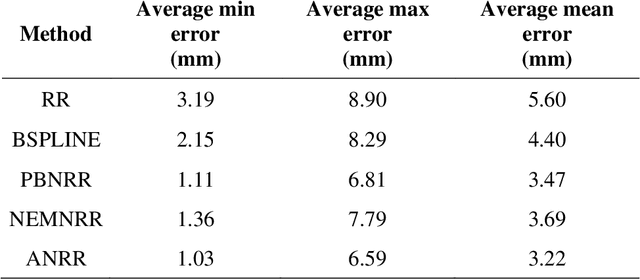
Abstract:Current neurosurgical procedures utilize medical images of various modalities to enable the precise location of tumors and critical brain structures to plan accurate brain tumor resection. The difficulty of using preoperative images during the surgery is caused by the intra-operative deformation of the brain tissue (brain shift), which introduces discrepancies concerning the preoperative configuration. Intra-operative imaging allows tracking such deformations but cannot fully substitute for the quality of the pre-operative data. Dynamic Data Driven Deformable Non-Rigid Registration (D4NRR) is a complex and time-consuming image processing operation that allows the dynamic adjustment of the pre-operative image data to account for intra-operative brain shift during the surgery. This paper summarizes the computational aspects of a specific adaptive numerical approximation method and its variations for registering brain MRIs. It outlines its evolution over the last 15 years and identifies new directions for the computational aspects of the technique.
Advancing Intra-operative Precision: Dynamic Data-Driven Non-Rigid Registration for Enhanced Brain Tumor Resection in Image-Guided Neurosurgery
Aug 31, 2023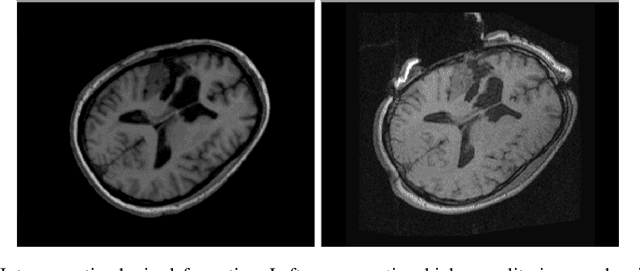
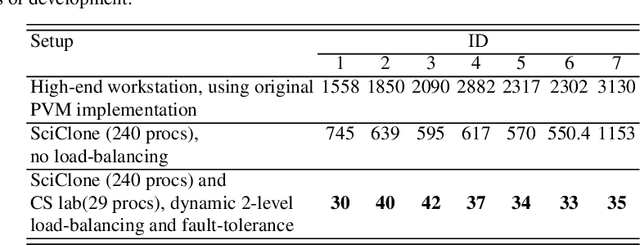
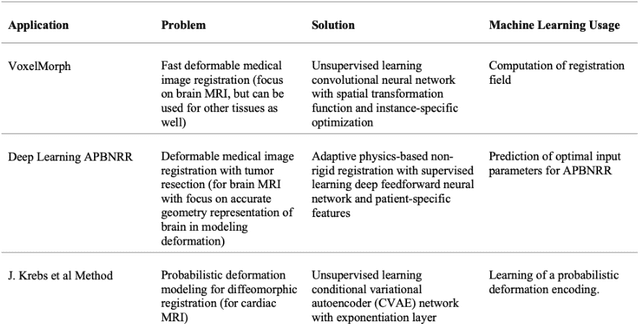
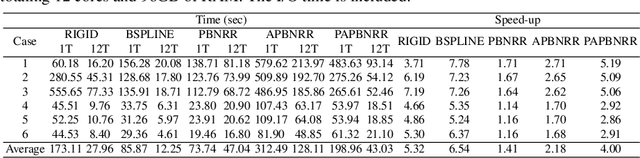
Abstract:During neurosurgery, medical images of the brain are used to locate tumors and critical structures, but brain tissue shifts make pre-operative images unreliable for accurate removal of tumors. Intra-operative imaging can track these deformations but is not a substitute for pre-operative data. To address this, we use Dynamic Data-Driven Non-Rigid Registration (NRR), a complex and time-consuming image processing operation that adjusts the pre-operative image data to account for intra-operative brain shift. Our review explores a specific NRR method for registering brain MRI during image-guided neurosurgery and examines various strategies for improving the accuracy and speed of the NRR method. We demonstrate that our implementation enables NRR results to be delivered within clinical time constraints while leveraging Distributed Computing and Machine Learning to enhance registration accuracy by identifying optimal parameters for the NRR method. Additionally, we highlight challenges associated with its use in the operating room.
Data Stealing Attack on Medical Images: Is it Safe to Export Networks from Data Lakes?
Jun 07, 2022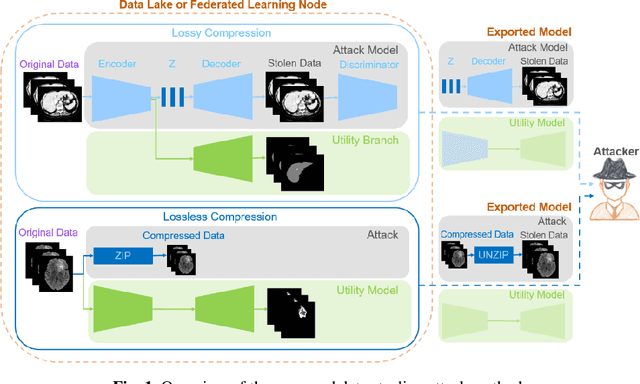
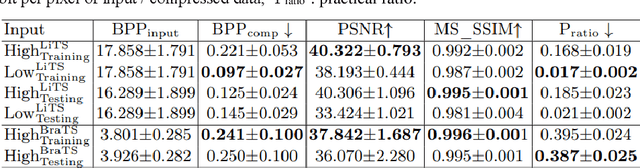
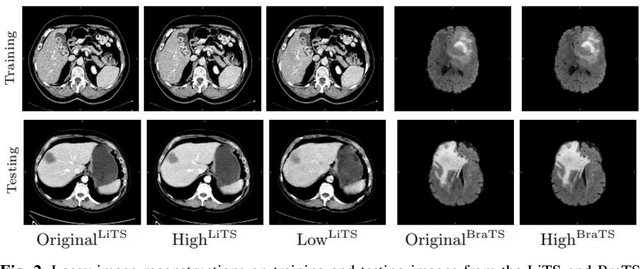
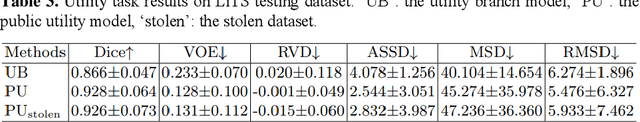
Abstract:In privacy-preserving machine learning, it is common that the owner of the learned model does not have any physical access to the data. Instead, only a secured remote access to a data lake is granted to the model owner without any ability to retrieve data from the data lake. Yet, the model owner may want to export the trained model periodically from the remote repository and a question arises whether this may cause is a risk of data leakage. In this paper, we introduce the concept of data stealing attack during the export of neural networks. It consists in hiding some information in the exported network that allows the reconstruction outside the data lake of images initially stored in that data lake. More precisely, we show that it is possible to train a network that can perform lossy image compression and at the same time solve some utility tasks such as image segmentation. The attack then proceeds by exporting the compression decoder network together with some image codes that leads to the image reconstruction outside the data lake. We explore the feasibility of such attacks on databases of CT and MR images, showing that it is possible to obtain perceptually meaningful reconstructions of the target dataset, and that the stolen dataset can be used in turns to solve a broad range of tasks. Comprehensive experiments and analyses show that data stealing attacks should be considered as a threat for sensitive imaging data sources.
Deep reinforcement learning in medical imaging: A literature review
Mar 05, 2021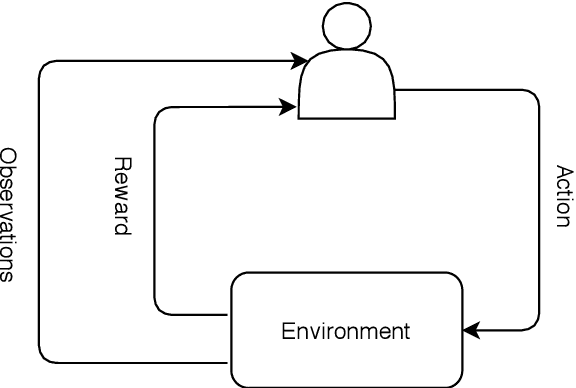
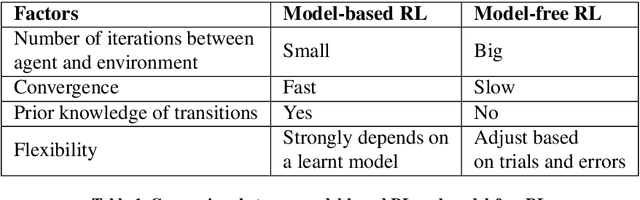

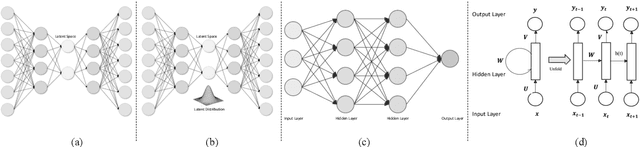
Abstract:Deep reinforcement learning (DRL) augments the reinforcement learning framework, which learns a sequence of actions that maximizes the expected reward, with the representative power of deep neural networks. Recent works have demonstrated the great potential of DRL in medicine and healthcare. This paper presents a literature review of DRL in medical imaging. We start with a comprehensive tutorial of DRL, including the latest model-free and model-based algorithms. We then cover existing DRL applications for medical imaging, which are roughly divided into three main categories: (I) parametric medical image analysis tasks including landmark detection, object/lesion detection, registration, and view plane localization; (ii) solving optimization tasks including hyperparameter tuning, selecting augmentation strategies, and neural architecture search; and (iii) miscellaneous applications including surgical gesture segmentation, personalized mobile health intervention, and computational model personalization. The paper concludes with discussions of future perspectives.
Combining Bayesian and Deep Learning Methods for the Delineation of the Fan in Ultrasound Images
Feb 02, 2021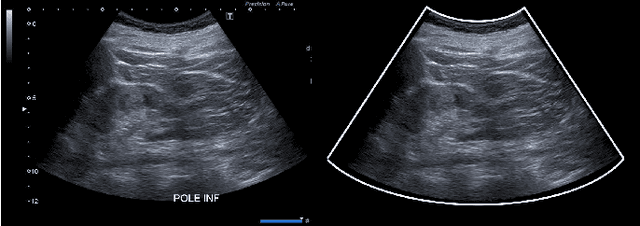
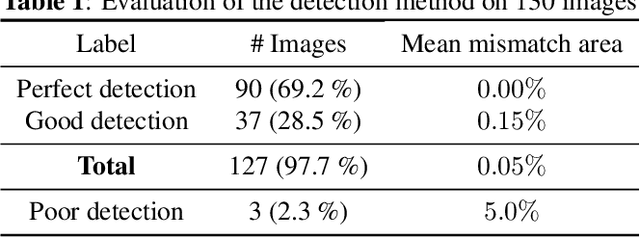
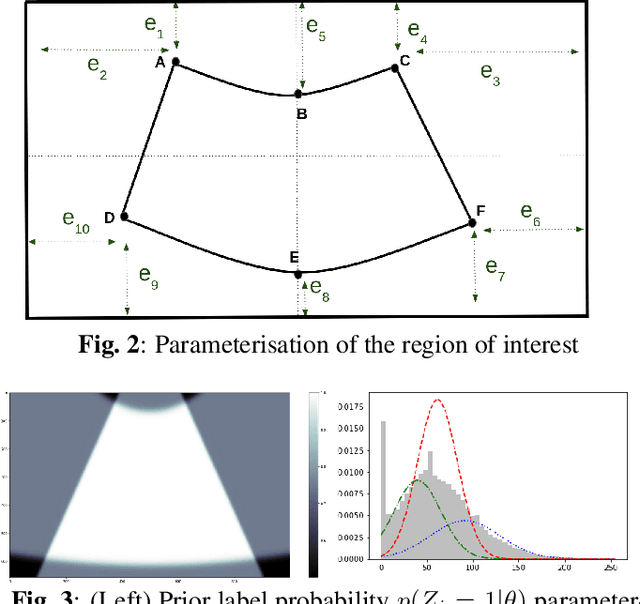
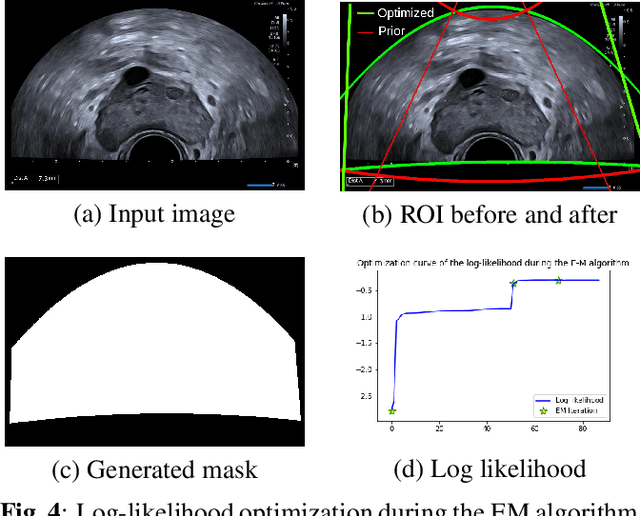
Abstract:Ultrasound (US) images usually contain identifying information outside the ultrasound fan area and manual annotations placed by the sonographers during exams. For those images to be exploitable in a Deep Learning framework, one needs to first delineate the border of the fan which delimits the ultrasound fan area and then remove other annotations inside. We propose a parametric probabilistic approach for the first task. We make use of this method to generate a training data set with segmentation masks of the region of interest (ROI) and train a U-Net to perform the same task in a supervised way, thus considerably reducing computational time of the method, one hundred and sixty times faster. These images are then processed with existing inpainting methods to remove annotations present inside the fan area. To the best of our knowledge, this is the first parametric approach to quickly detect the fan in an ultrasound image without any other information than the image itself.
Learning a Generative Motion Model from Image Sequences based on a Latent Motion Matrix
Nov 03, 2020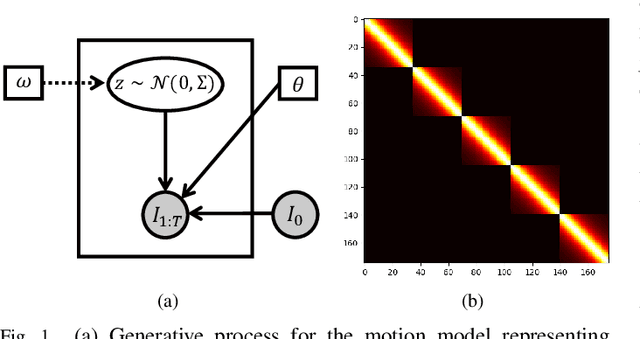
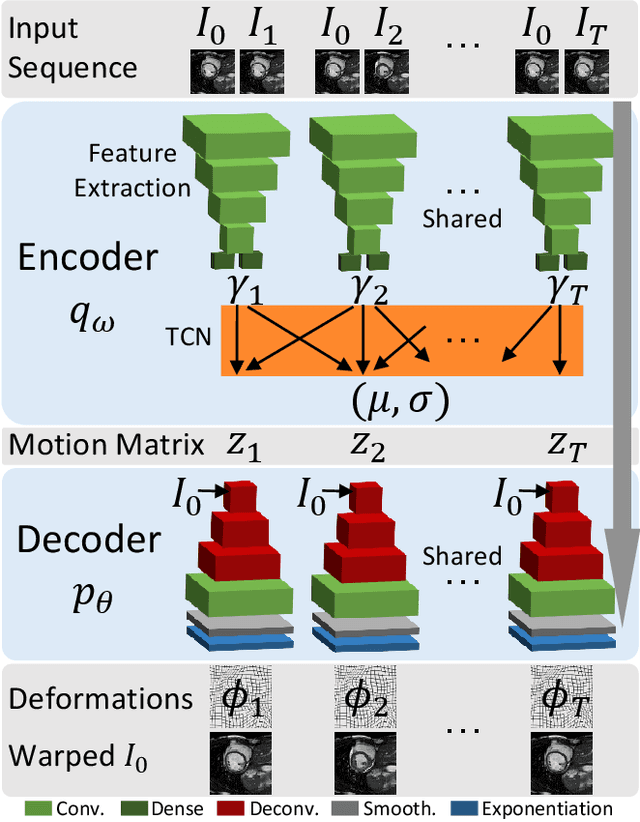
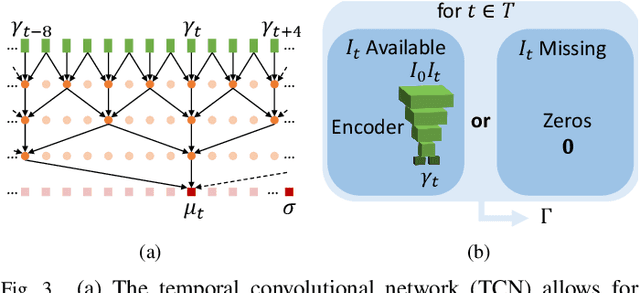
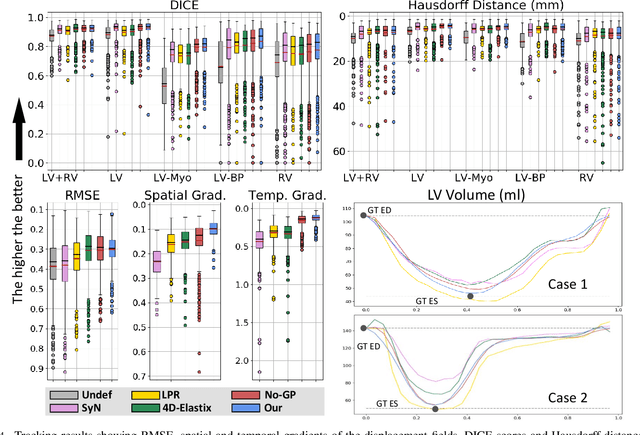
Abstract:We propose to learn a probabilistic motion model from a sequence of images for spatio-temporal registration. Our model encodes motion in a low-dimensional probabilistic space - the motion matrix - which enables various motion analysis tasks such as simulation and interpolation of realistic motion patterns allowing for faster data acquisition and data augmentation. More precisely, the motion matrix allows to transport the recovered motion from one subject to another simulating for example a pathological motion in a healthy subject without the need for inter-subject registration. The method is based on a conditional latent variable model that is trained using amortized variational inference. This unsupervised generative model follows a novel multivariate Gaussian process prior and is applied within a temporal convolutional network which leads to a diffeomorphic motion model. Temporal consistency and generalizability is further improved by applying a temporal dropout training scheme. Applied to cardiac cine-MRI sequences, we show improved registration accuracy and spatio-temporally smoother deformations compared to three state-of-the-art registration algorithms. Besides, we demonstrate the model's applicability for motion analysis, simulation and super-resolution by an improved motion reconstruction from sequences with missing frames compared to linear and cubic interpolation.
Probabilistic Motion Modeling from Medical Image Sequences: Application to Cardiac Cine-MRI
Jul 31, 2019

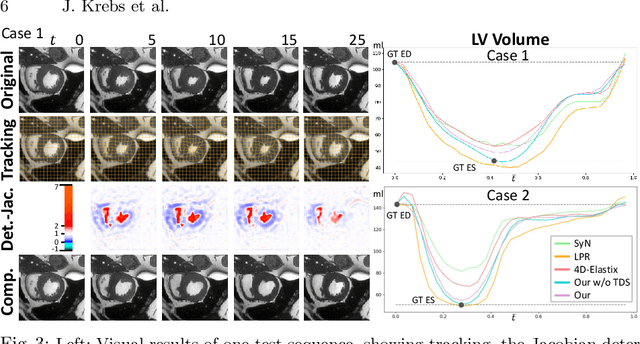

Abstract:We propose to learn a probabilistic motion model from a sequence of images. Besides spatio-temporal registration, our method offers to predict motion from a limited number of frames, useful for temporal super-resolution. The model is based on a probabilistic latent space and a novel temporal dropout training scheme. This enables simulation and interpolation of realistic motion patterns given only one or any subset of frames of a sequence. The encoded motion also allows to be transported from one subject to another without the need of inter-subject registration. An unsupervised generative deformation model is applied within a temporal convolutional network which leads to a diffeomorphic motion model, encoded as a low-dimensional motion matrix. Applied to cardiac cine-MRI sequences, we show improved registration accuracy and spatio-temporally smoother deformations compared to three state-of-the-art registration algorithms. Besides, we demonstrate the model's applicability to motion transport by simulating a pathology in a healthy case. Furthermore, we show an improved motion reconstruction from incomplete sequences compared to linear and cubic interpolation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge