Julian Krebs
Learning a Generative Motion Model from Image Sequences based on a Latent Motion Matrix
Nov 03, 2020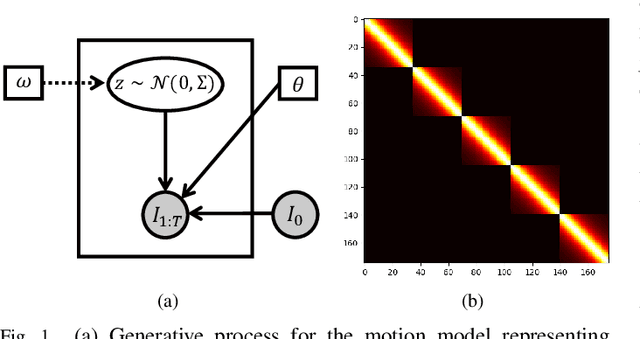
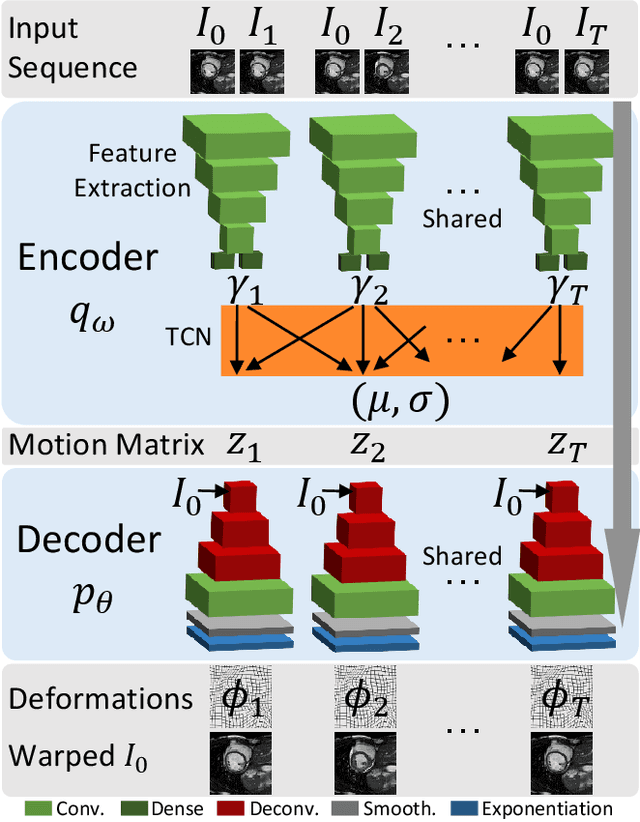
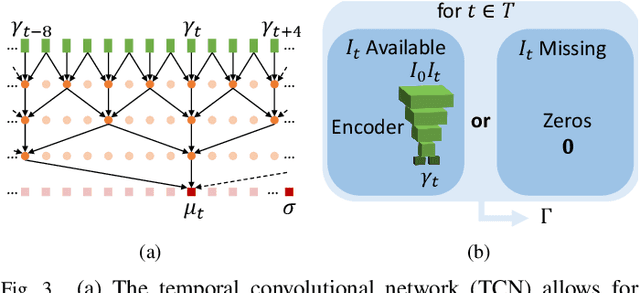
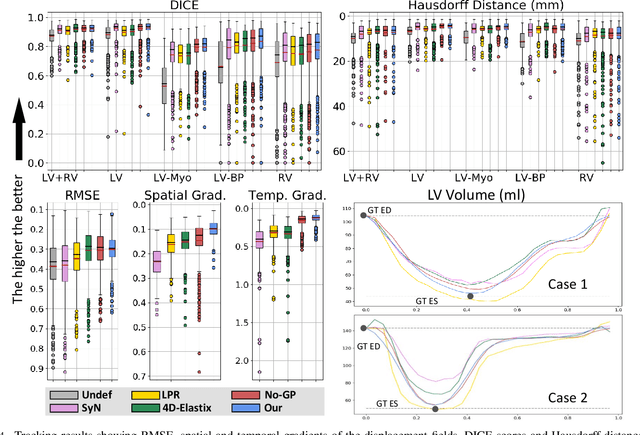
Abstract:We propose to learn a probabilistic motion model from a sequence of images for spatio-temporal registration. Our model encodes motion in a low-dimensional probabilistic space - the motion matrix - which enables various motion analysis tasks such as simulation and interpolation of realistic motion patterns allowing for faster data acquisition and data augmentation. More precisely, the motion matrix allows to transport the recovered motion from one subject to another simulating for example a pathological motion in a healthy subject without the need for inter-subject registration. The method is based on a conditional latent variable model that is trained using amortized variational inference. This unsupervised generative model follows a novel multivariate Gaussian process prior and is applied within a temporal convolutional network which leads to a diffeomorphic motion model. Temporal consistency and generalizability is further improved by applying a temporal dropout training scheme. Applied to cardiac cine-MRI sequences, we show improved registration accuracy and spatio-temporally smoother deformations compared to three state-of-the-art registration algorithms. Besides, we demonstrate the model's applicability for motion analysis, simulation and super-resolution by an improved motion reconstruction from sequences with missing frames compared to linear and cubic interpolation.
Probabilistic Motion Modeling from Medical Image Sequences: Application to Cardiac Cine-MRI
Jul 31, 2019

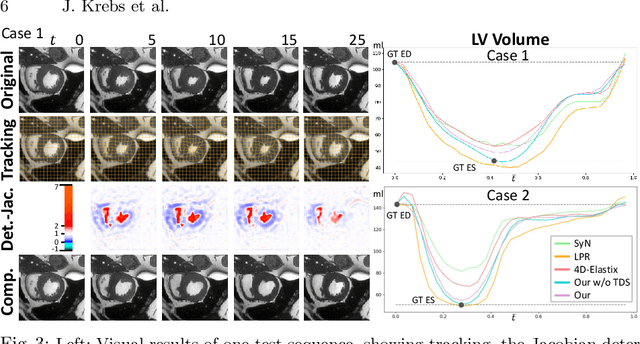

Abstract:We propose to learn a probabilistic motion model from a sequence of images. Besides spatio-temporal registration, our method offers to predict motion from a limited number of frames, useful for temporal super-resolution. The model is based on a probabilistic latent space and a novel temporal dropout training scheme. This enables simulation and interpolation of realistic motion patterns given only one or any subset of frames of a sequence. The encoded motion also allows to be transported from one subject to another without the need of inter-subject registration. An unsupervised generative deformation model is applied within a temporal convolutional network which leads to a diffeomorphic motion model, encoded as a low-dimensional motion matrix. Applied to cardiac cine-MRI sequences, we show improved registration accuracy and spatio-temporally smoother deformations compared to three state-of-the-art registration algorithms. Besides, we demonstrate the model's applicability to motion transport by simulating a pathology in a healthy case. Furthermore, we show an improved motion reconstruction from incomplete sequences compared to linear and cubic interpolation.
Learning a Probabilistic Model for Diffeomorphic Registration
Dec 18, 2018



Abstract:We propose to learn a low-dimensional probabilistic deformation model from data which can be used for registration and the analysis of deformations. The latent variable model maps similar deformations close to each other in an encoding space. It enables to compare deformations, generate normal or pathological deformations for any new image or to transport deformations from one image pair to any other image. Our unsupervised method is based on variational inference. In particular, we use a conditional variational autoencoder (CVAE) network and constrain transformations to be symmetric and diffeomorphic by applying a differentiable exponentiation layer with a symmetric loss function. We also present a formulation that includes spatial regularization such as diffusion-based filters. Additionally, our framework provides multi-scale velocity field estimations. We evaluated our method on 3-D intra-subject registration using 334 cardiac cine-MRIs. On this dataset, our method showed state-of-the-art performance with a mean DICE score of 81.2% and a mean Hausdorff distance of 7.3mm using 32 latent dimensions compared to three state-of-the-art methods while also demonstrating more regular deformation fields. The average time per registration was 0.32s. Besides, we visualized the learned latent space and show that the encoded deformations can be used to transport deformations and to cluster diseases with a classification accuracy of 83% after applying a linear projection.
Unsupervised Probabilistic Deformation Modeling for Robust Diffeomorphic Registration
Jul 20, 2018



Abstract:We propose a deformable registration algorithm based on unsupervised learning of a low-dimensional probabilistic parameterization of deformations. We model registration in a probabilistic and generative fashion, by applying a conditional variational autoencoder (CVAE) network. This model enables to also generate normal or pathological deformations of any new image based on the probabilistic latent space. Most recent learning-based registration algorithms use supervised labels or deformation models, that miss important properties such as diffeomorphism and sufficiently regular deformation fields. In this work, we constrain transformations to be diffeomorphic by using a differentiable exponentiation layer with a symmetric loss function. We evaluated our method on 330 cardiac MR sequences and demonstrate robust intra-subject registration results comparable to two state-of-the-art methods but with more regular deformation fields compared to a recent learning-based algorithm. Our method reached a mean DICE score of 78.3% and a mean Hausdorff distance of 7.9mm. In two preliminary experiments, we illustrate the model's abilities to transport pathological deformations to healthy subjects and to cluster five diseases in the unsupervised deformation encoding space with a classification performance of 70%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge