P. Ellen Grant
Atlas-Assisted Segment Anything Model for Fetal Brain MRI (FeTal-SAM)
Jan 22, 2026Abstract:This paper presents FeTal-SAM, a novel adaptation of the Segment Anything Model (SAM) tailored for fetal brain MRI segmentation. Traditional deep learning methods often require large annotated datasets for a fixed set of labels, making them inflexible when clinical or research needs change. By integrating atlas-based prompts and foundation-model principles, FeTal-SAM addresses two key limitations in fetal brain MRI segmentation: (1) the need to retrain models for varying label definitions, and (2) the lack of insight into whether segmentations are driven by genuine image contrast or by learned spatial priors. We leverage multi-atlas registration to generate spatially aligned label templates that serve as dense prompts, alongside a bounding-box prompt, for SAM's segmentation decoder. This strategy enables binary segmentation on a per-structure basis, which is subsequently fused to reconstruct the full 3D segmentation volumes. Evaluations on two datasets, the dHCP dataset and an in-house dataset demonstrate FeTal-SAM's robust performance across gestational ages. Notably, it achieves Dice scores comparable to state-of-the-art baselines which were trained for each dataset and label definition for well-contrasted structures like cortical plate and cerebellum, while maintaining the flexibility to segment any user-specified anatomy. Although slightly lower accuracy is observed for subtle, low-contrast structures (e.g., hippocampus, amygdala), our results highlight FeTal-SAM's potential to serve as a general-purpose segmentation model without exhaustive retraining. This method thus constitutes a promising step toward clinically adaptable fetal brain MRI analysis tools.
Fast Multi-Stack Slice-to-Volume Reconstruction via Multi-Scale Unrolled Optimization
Jan 12, 2026Abstract:Fully convolutional networks have become the backbone of modern medical imaging due to their ability to learn multi-scale representations and perform end-to-end inference. Yet their potential for slice-to-volume reconstruction (SVR), the task of jointly estimating 3D anatomy and slice poses from misaligned 2D acquisitions, remains underexplored. We introduce a fast convolutional framework that fuses multiple orthogonal 2D slice stacks to recover coherent 3D structure and refines slice alignment through lightweight model-based optimization. Applied to fetal brain MRI, our approach reconstructs high-quality 3D volumes in under 10s, with 1s slice registration and accuracy on par with state-of-the-art iterative SVR pipelines, offering more than speedup. The framework uses non-rigid displacement fields to represent transformations, generalizing to other SVR problems like fetal body and placental MRI. Additionally, the fast inference time paves the way for real-time, scanner-side volumetric feedback during MRI acquisition.
USFetal: Tools for Fetal Brain Ultrasound Compounding
Jan 11, 2026Abstract:Ultrasound offers a safe, cost-effective, and widely accessible technology for fetal brain imaging, making it especially suitable for routine clinical use. However, it suffers from view-dependent artifacts, operator variability, and a limited field of view, which make interpretation and quantitative evaluation challenging. Ultrasound compounding aims to overcome these limitations by integrating complementary information from multiple 3D acquisitions into a single, coherent volumetric representation. This work provides four main contributions: (1) We present the first systematic categorization of computational strategies for fetal brain ultrasound compounding, including both classical techniques and modern learning-based frameworks. (2) We implement and compare representative methods across four key categories - multi-scale, transformation-based, variational, and deep learning approaches - emphasizing their core principles and practical advantages. (3) Motivated by the lack of full-view, artifact-free ground truth required for supervised learning, we focus on unsupervised and self-supervised strategies and introduce two new deep learning based approaches: a self-supervised compounding framework and an adaptation of unsupervised deep plug-and-play priors for compounding. (4) We conduct a comprehensive evaluation on ten multi-view fetal brain ultrasound datasets, using both expert radiologist scoring and standard quantitative image-quality metrics. We also release the USFetal Compounding Toolbox, publicly available to support benchmarking and future research. Keywords: Ultrasound compounding, fetal brain, deep learning, self-supervised, unsupervised.
Spatial regularisation for improved accuracy and interpretability in keypoint-based registration
Mar 07, 2025Abstract:Unsupervised registration strategies bypass requirements in ground truth transforms or segmentations by optimising similarity metrics between fixed and moved volumes. Among these methods, a recent subclass of approaches based on unsupervised keypoint detection stand out as very promising for interpretability. Specifically, these methods train a network to predict feature maps for fixed and moving images, from which explainable centres of mass are computed to obtain point clouds, that are then aligned in closed-form. However, the features returned by the network often yield spatially diffuse patterns that are hard to interpret, thus undermining the purpose of keypoint-based registration. Here, we propose a three-fold loss to regularise the spatial distribution of the features. First, we use the KL divergence to model features as point spread functions that we interpret as probabilistic keypoints. Then, we sharpen the spatial distributions of these features to increase the precision of the detected landmarks. Finally, we introduce a new repulsive loss across keypoints to encourage spatial diversity. Overall, our loss considerably improves the interpretability of the features, which now correspond to precise and anatomically meaningful landmarks. We demonstrate our three-fold loss in foetal rigid motion tracking and brain MRI affine registration tasks, where it not only outperforms state-of-the-art unsupervised strategies, but also bridges the gap with state-of-the-art supervised methods. Our code is available at https://github.com/BenBillot/spatial_regularisation.
Relation U-Net
Jan 15, 2025Abstract:Towards clinical interpretations, this paper presents a new ''output-with-confidence'' segmentation neural network with multiple input images and multiple output segmentation maps and their pairwise relations. A confidence score of the test image without ground-truth can be estimated from the difference among the estimated relation maps. We evaluate the method based on the widely used vanilla U-Net for segmentation and our new model is named Relation U-Net which can output segmentation maps of the input images as well as an estimated confidence score of the test image without ground-truth. Experimental results on four public datasets show that Relation U-Net can not only provide better accuracy than vanilla U-Net but also estimate a confidence score which is linearly correlated to the segmentation accuracy on test images.
Learning General-Purpose Biomedical Volume Representations using Randomized Synthesis
Nov 04, 2024Abstract:Current volumetric biomedical foundation models struggle to generalize as public 3D datasets are small and do not cover the broad diversity of medical procedures, conditions, anatomical regions, and imaging protocols. We address this by creating a representation learning method that instead anticipates strong domain shifts at training time itself. We first propose a data engine that synthesizes highly variable training samples that enable generalization to new biomedical contexts. To then train a single 3D network for any voxel-level task, we develop a contrastive learning method that pretrains the network to be stable against nuisance imaging variation simulated by the data engine, a key inductive bias for generalization. This network's features can be used as robust representations of input images for downstream tasks and its weights provide a strong, dataset-agnostic initialization for finetuning on new datasets. As a result, we set new standards across both multimodality registration and few-shot segmentation, a first for any 3D biomedical vision model, all without (pre-)training on any existing dataset of real images.
Shape-aware Segmentation of the Placenta in BOLD Fetal MRI Time Series
Dec 08, 2023


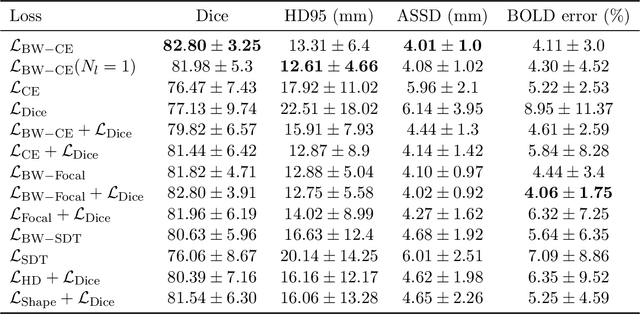
Abstract:Blood oxygen level dependent (BOLD) MRI time series with maternal hyperoxia can assess placental oxygenation and function. Measuring precise BOLD changes in the placenta requires accurate temporal placental segmentation and is confounded by fetal and maternal motion, contractions, and hyperoxia-induced intensity changes. Current BOLD placenta segmentation methods warp a manually annotated subject-specific template to the entire time series. However, as the placenta is a thin, elongated, and highly non-rigid organ subject to large deformations and obfuscated edges, existing work cannot accurately segment the placental shape, especially near boundaries. In this work, we propose a machine learning segmentation framework for placental BOLD MRI and apply it to segmenting each volume in a time series. We use a placental-boundary weighted loss formulation and perform a comprehensive evaluation across several popular segmentation objectives. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. Biomedically, our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. We further find that boundary-weighting increases placental segmentation performance by 8.3% and 6.0% Dice coefficient for the cross-entropy and signed distance transform objectives, respectively. Our code and trained model is available at https://github.com/mabulnaga/automatic-placenta-segmentation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:017. arXiv admin note: substantial text overlap with arXiv:2208.02895
Dynamic Neural Fields for Learning Atlases of 4D Fetal MRI Time-series
Nov 06, 2023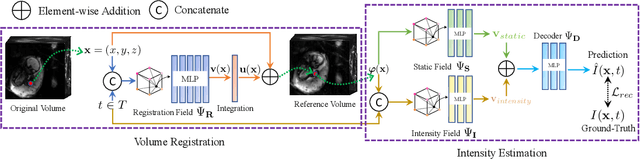
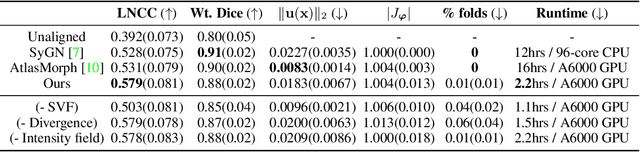
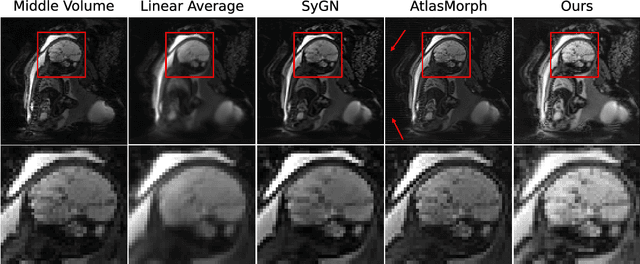
Abstract:We present a method for fast biomedical image atlas construction using neural fields. Atlases are key to biomedical image analysis tasks, yet conventional and deep network estimation methods remain time-intensive. In this preliminary work, we frame subject-specific atlas building as learning a neural field of deformable spatiotemporal observations. We apply our method to learning subject-specific atlases and motion stabilization of dynamic BOLD MRI time-series of fetuses in utero. Our method yields high-quality atlases of fetal BOLD time-series with $\sim$5-7$\times$ faster convergence compared to existing work. While our method slightly underperforms well-tuned baselines in terms of anatomical overlap, it estimates templates significantly faster, thus enabling rapid processing and stabilization of large databases of 4D dynamic MRI acquisitions. Code is available at https://github.com/Kidrauh/neural-atlasing
Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series
Oct 16, 2023Abstract:The placenta plays a crucial role in fetal development. Automated 3D placenta segmentation from fetal EPI MRI holds promise for advancing prenatal care. This paper proposes an effective semi-supervised learning method for improving placenta segmentation in fetal EPI MRI time series. We employ consistency regularization loss that promotes consistency under spatial transformation of the same image and temporal consistency across nearby images in a time series. The experimental results show that the method improves the overall segmentation accuracy and provides better performance for outliers and hard samples. The evaluation also indicates that our method improves the temporal coherency of the prediction, which could lead to more accurate computation of temporal placental biomarkers. This work contributes to the study of the placenta and prenatal clinical decision-making. Code is available at https://github.com/firstmover/cr-seg.
AnyStar: Domain randomized universal star-convex 3D instance segmentation
Jul 13, 2023Abstract:Star-convex shapes arise across bio-microscopy and radiology in the form of nuclei, nodules, metastases, and other units. Existing instance segmentation networks for such structures train on densely labeled instances for each dataset, which requires substantial and often impractical manual annotation effort. Further, significant reengineering or finetuning is needed when presented with new datasets and imaging modalities due to changes in contrast, shape, orientation, resolution, and density. We present AnyStar, a domain-randomized generative model that simulates synthetic training data of blob-like objects with randomized appearance, environments, and imaging physics to train general-purpose star-convex instance segmentation networks. As a result, networks trained using our generative model do not require annotated images from unseen datasets. A single network trained on our synthesized data accurately 3D segments C. elegans and P. dumerilii nuclei in fluorescence microscopy, mouse cortical nuclei in micro-CT, zebrafish brain nuclei in EM, and placental cotyledons in human fetal MRI, all without any retraining, finetuning, transfer learning, or domain adaptation. Code is available at https://github.com/neel-dey/AnyStar.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge