Yangming Ou
Relation U-Net
Jan 15, 2025Abstract:Towards clinical interpretations, this paper presents a new ''output-with-confidence'' segmentation neural network with multiple input images and multiple output segmentation maps and their pairwise relations. A confidence score of the test image without ground-truth can be estimated from the difference among the estimated relation maps. We evaluate the method based on the widely used vanilla U-Net for segmentation and our new model is named Relation U-Net which can output segmentation maps of the input images as well as an estimated confidence score of the test image without ground-truth. Experimental results on four public datasets show that Relation U-Net can not only provide better accuracy than vanilla U-Net but also estimate a confidence score which is linearly correlated to the segmentation accuracy on test images.
AGE2HIE: Transfer Learning from Brain Age to Predicting Neurocognitive Outcome for Infant Brain Injury
Nov 07, 2024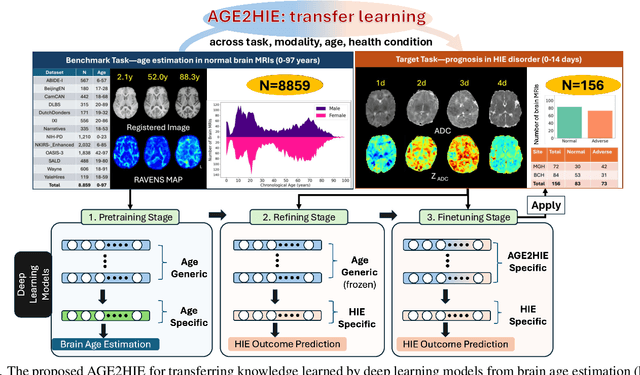
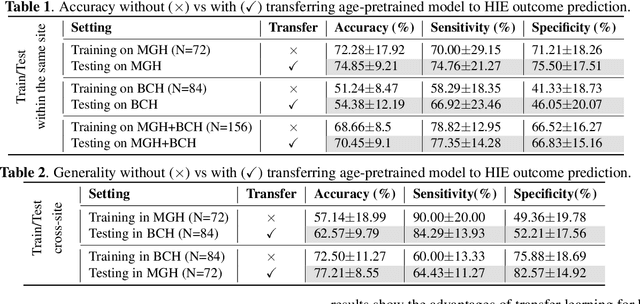
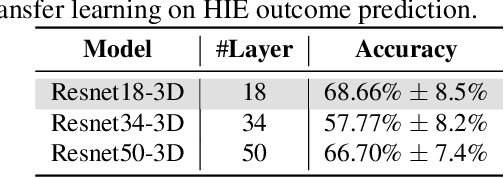
Abstract:Hypoxic-Ischemic Encephalopathy (HIE) affects 1 to 5 out of every 1,000 newborns, with 30% to 50% of cases resulting in adverse neurocognitive outcomes. However, these outcomes can only be reliably assessed as early as age 2. Therefore, early and accurate prediction of HIE-related neurocognitive outcomes using deep learning models is critical for improving clinical decision-making, guiding treatment decisions and assessing novel therapies. However, a major challenge in developing deep learning models for this purpose is the scarcity of large, annotated HIE datasets. We have assembled the first and largest public dataset, however it contains only 156 cases with 2-year neurocognitive outcome labels. In contrast, we have collected 8,859 normal brain black Magnetic Resonance Imagings (MRIs) with 0-97 years of age that are available for brain age estimation using deep learning models. In this paper, we introduce AGE2HIE to transfer knowledge learned by deep learning models from healthy controls brain MRIs to a diseased cohort, from structural to diffusion MRIs, from regression of continuous age estimation to prediction of the binary neurocognitive outcomes, and from lifespan age (0-97 years) to infant (0-2 weeks). Compared to training from scratch, transfer learning from brain age estimation significantly improves not only the prediction accuracy (3% or 2% improvement in same or multi-site), but also the model generalization across different sites (5% improvement in cross-site validation).
BOston Neonatal Brain Injury Data for Hypoxic Ischemic Encephalopathy (BONBID-HIE): II. 2-year Neurocognitive Outcome and NICU Outcome
Nov 05, 2024


Abstract:Hypoxic Ischemic Encephalopathy (HIE) affects approximately 1-5/1000 newborns globally and leads to adverse neurocognitive outcomes in 30% to 50% of cases by two years of age. Despite therapeutic advances with Therapeutic Hypothermia (TH), prognosis remains challenging, highlighting the need for improved biomarkers. This paper introduces the second release of the Boston Neonatal Brain Injury Dataset for Hypoxic-Ischemic Encephalopathy (BONBID-HIE), an open-source, comprehensive MRI and clinical dataset featuring 237 patients, including NICU outcomes and 2-year neurocognitive outcomes from Massachusetts General Hospital and Boston Children's Hospital.
Foundation AI Model for Medical Image Segmentation
Nov 05, 2024Abstract:Foundation models refer to artificial intelligence (AI) models that are trained on massive amounts of data and demonstrate broad generalizability across various tasks with high accuracy. These models offer versatile, one-for-many or one-for-all solutions, eliminating the need for developing task-specific AI models. Examples of such foundation models include the Chat Generative Pre-trained Transformer (ChatGPT) and the Segment Anything Model (SAM). These models have been trained on millions to billions of samples and have shown wide-ranging and accurate applications in numerous tasks such as text processing (using ChatGPT) and natural image segmentation (using SAM). In medical image segmentation - finding target regions in medical images - there is a growing need for these one-for-many or one-for-all foundation models. Such models could obviate the need to develop thousands of task-specific AI models, which is currently standard practice in the field. They can also be adapted to tasks with datasets too small for effective training. We discuss two paths to achieve foundation models for medical image segmentation and comment on progress, challenges, and opportunities. One path is to adapt or fine-tune existing models, originally developed for natural images, for use with medical images. The second path entails building models from scratch, exclusively training on medical images.
A Self-Supervised Image Registration Approach for Measuring Local Response Patterns in Metastatic Ovarian Cancer
Jul 24, 2024



Abstract:High-grade serous ovarian carcinoma (HGSOC) is characterised by significant spatial and temporal heterogeneity, typically manifesting at an advanced metastatic stage. A major challenge in treating advanced HGSOC is effectively monitoring localised change in tumour burden across multiple sites during neoadjuvant chemotherapy (NACT) and predicting long-term pathological response and overall patient survival. In this work, we propose a self-supervised deformable image registration algorithm that utilises a general-purpose image encoder for image feature extraction to co-register contrast-enhanced computerised tomography scan images acquired before and after neoadjuvant chemotherapy. This approach addresses challenges posed by highly complex tumour deformations and longitudinal lesion matching during treatment. Localised tumour changes are calculated using the Jacobian determinant maps of the registration deformation at multiple disease sites and their macroscopic areas, including hypo-dense (i.e., cystic/necrotic), hyper-dense (i.e., calcified), and intermediate density (i.e., soft tissue) portions. A series of experiments is conducted to understand the role of a general-purpose image encoder and its application in quantifying change in tumour burden during neoadjuvant chemotherapy in HGSOC. This work is the first to demonstrate the feasibility of a self-supervised image registration approach in quantifying NACT-induced localised tumour changes across the whole disease burden of patients with complex multi-site HGSOC, which could be used as a potential marker for ovarian cancer patient's long-term pathological response and survival.
Accuracy of Segment-Anything Model in medical image segmentation tasks
Apr 27, 2023Abstract:The segment-anything model (SAM), was introduced as a fundamental model for segmenting images. It was trained using over 1 billion masks from 11 million natural images. The model can perform zero-shot segmentation of images by using various prompts such as masks, boxes, and points. In this report, we explored (1) the accuracy of SAM on 12 public medical image segmentation datasets which cover various organs (brain, breast, chest, lung, skin, liver, bowel, pancreas, and prostate), image modalities (2D X-ray, histology, endoscropy, and 3D MRI and CT), and health conditions (normal, lesioned). (2) if the computer vision foundational segmentation model SAM can provide promising research directions for medical image segmentation. We found that SAM without re-training on medical images does not perform as accurately as U-Net or other deep learning models trained on medical images.
U-Netmer: U-Net meets Transformer for medical image segmentation
Apr 03, 2023Abstract:The combination of the U-Net based deep learning models and Transformer is a new trend for medical image segmentation. U-Net can extract the detailed local semantic and texture information and Transformer can learn the long-rang dependencies among pixels in the input image. However, directly adapting the Transformer for segmentation has ``token-flatten" problem (flattens the local patches into 1D tokens which losses the interaction among pixels within local patches) and ``scale-sensitivity" problem (uses a fixed scale to split the input image into local patches). Compared to directly combining U-Net and Transformer, we propose a new global-local fashion combination of U-Net and Transformer, named U-Netmer, to solve the two problems. The proposed U-Netmer splits an input image into local patches. The global-context information among local patches is learnt by the self-attention mechanism in Transformer and U-Net segments each local patch instead of flattening into tokens to solve the `token-flatten" problem. The U-Netmer can segment the input image with different patch sizes with the identical structure and the same parameter. Thus, the U-Netmer can be trained with different patch sizes to solve the ``scale-sensitivity" problem. We conduct extensive experiments in 7 public datasets on 7 organs (brain, heart, breast, lung, polyp, pancreas and prostate) and 4 imaging modalities (MRI, CT, ultrasound, and endoscopy) to show that the proposed U-Netmer can be generally applied to improve accuracy of medical image segmentation. These experimental results show that U-Netmer provides state-of-the-art performance compared to baselines and other models. In addition, the discrepancy among the outputs of U-Netmer with different scales is linearly correlated to the segmentation accuracy which can be considered as a confidence score to rank test images by difficulty without ground-truth.
Segmentation Ability Map: Interpret deep features for medical image segmentation
Dec 19, 2022Abstract:Deep convolutional neural networks (CNNs) have been widely used for medical image segmentation. In most studies, only the output layer is exploited to compute the final segmentation results and the hidden representations of the deep learned features have not been well understood. In this paper, we propose a prototype segmentation (ProtoSeg) method to compute a binary segmentation map based on deep features. We measure the segmentation abilities of the features by computing the Dice between the feature segmentation map and ground-truth, named as the segmentation ability score (SA score for short). The corresponding SA score can quantify the segmentation abilities of deep features in different layers and units to understand the deep neural networks for segmentation. In addition, our method can provide a mean SA score which can give a performance estimation of the output on the test images without ground-truth. Finally, we use the proposed ProtoSeg method to compute the segmentation map directly on input images to further understand the segmentation ability of each input image. Results are presented on segmenting tumors in brain MRI, lesions in skin images, COVID-related abnormality in CT images, prostate segmentation in abdominal MRI, and pancreatic mass segmentation in CT images. Our method can provide new insights for interpreting and explainable AI systems for medical image segmentation. Our code is available on: \url{https://github.com/shengfly/ProtoSeg}.
Deep Relation Learning for Regression and Its Application to Brain Age Estimation
Apr 13, 2022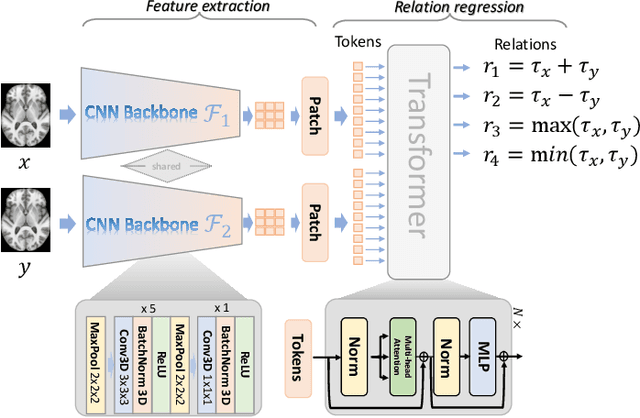
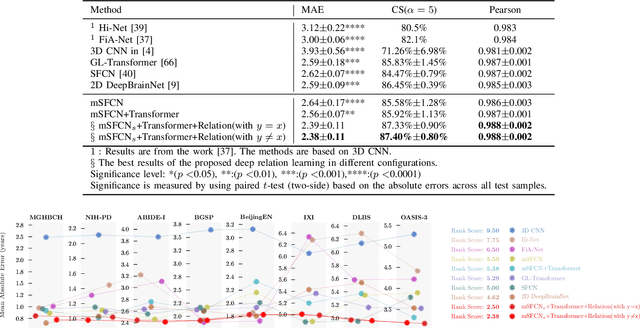
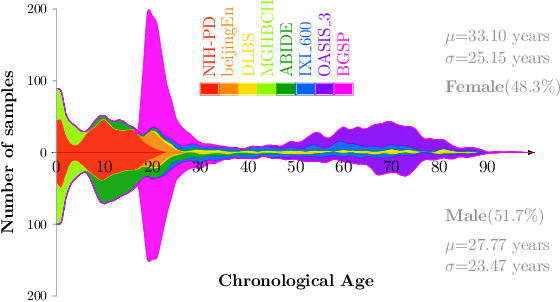
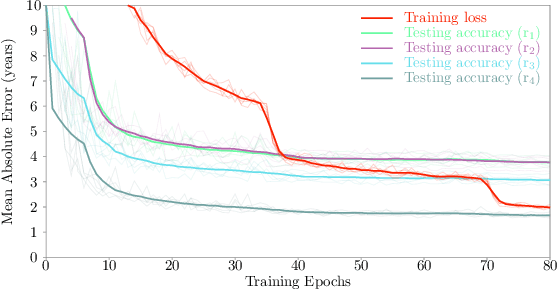
Abstract:Most deep learning models for temporal regression directly output the estimation based on single input images, ignoring the relationships between different images. In this paper, we propose deep relation learning for regression, aiming to learn different relations between a pair of input images. Four non-linear relations are considered: "cumulative relation", "relative relation", "maximal relation" and "minimal relation". These four relations are learned simultaneously from one deep neural network which has two parts: feature extraction and relation regression. We use an efficient convolutional neural network to extract deep features from the pair of input images and apply a Transformer for relation learning. The proposed method is evaluated on a merged dataset with 6,049 subjects with ages of 0-97 years using 5-fold cross-validation for the task of brain age estimation. The experimental results have shown that the proposed method achieved a mean absolute error (MAE) of 2.38 years, which is lower than the MAEs of 8 other state-of-the-art algorithms with statistical significance (p$<$0.05) in paired T-test (two-side).
Global-Local Transformer for Brain Age Estimation
Sep 03, 2021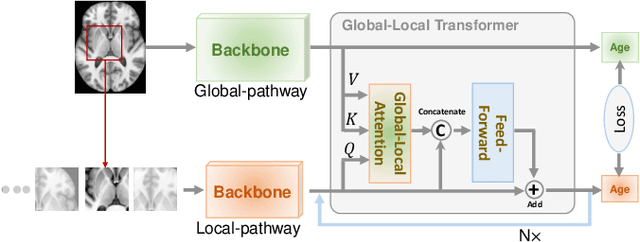
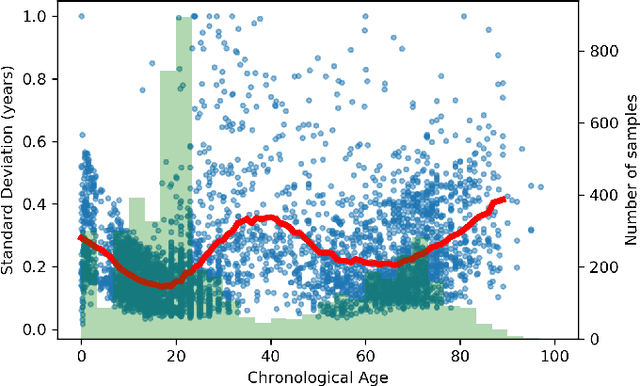
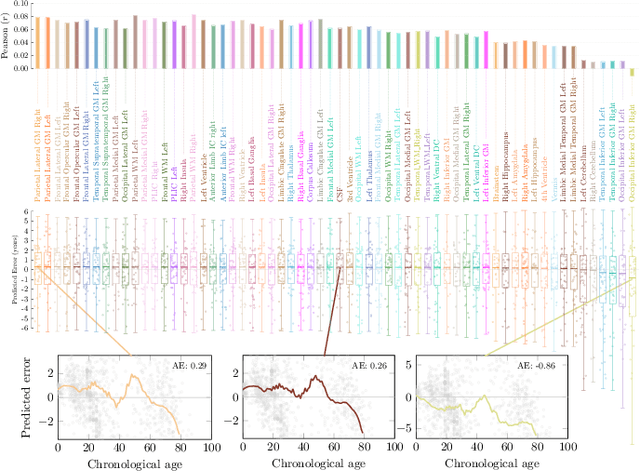
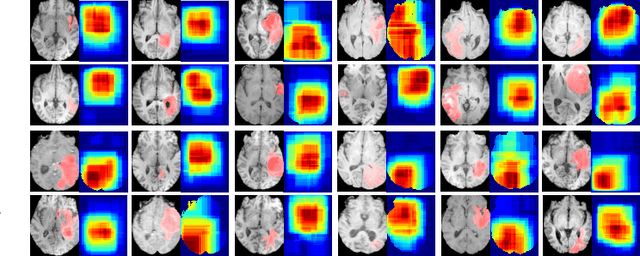
Abstract:Deep learning can provide rapid brain age estimation based on brain magnetic resonance imaging (MRI). However, most studies use one neural network to extract the global information from the whole input image, ignoring the local fine-grained details. In this paper, we propose a global-local transformer, which consists of a global-pathway to extract the global-context information from the whole input image and a local-pathway to extract the local fine-grained details from local patches. The fine-grained information from the local patches are fused with the global-context information by the attention mechanism, inspired by the transformer, to estimate the brain age. We evaluate the proposed method on 8 public datasets with 8,379 healthy brain MRIs with the age range of 0-97 years. 6 datasets are used for cross-validation and 2 datasets are used for evaluating the generality. Comparing with other state-of-the-art methods, the proposed global-local transformer reduces the mean absolute error of the estimated ages to 2.70 years and increases the correlation coefficient of the estimated age and the chronological age to 0.9853. In addition, our proposed method provides regional information of which local patches are most informative for brain age estimation. Our source code is available on: \url{https://github.com/shengfly/global-local-transformer}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge