Rina Bao
Relation U-Net
Jan 15, 2025Abstract:Towards clinical interpretations, this paper presents a new ''output-with-confidence'' segmentation neural network with multiple input images and multiple output segmentation maps and their pairwise relations. A confidence score of the test image without ground-truth can be estimated from the difference among the estimated relation maps. We evaluate the method based on the widely used vanilla U-Net for segmentation and our new model is named Relation U-Net which can output segmentation maps of the input images as well as an estimated confidence score of the test image without ground-truth. Experimental results on four public datasets show that Relation U-Net can not only provide better accuracy than vanilla U-Net but also estimate a confidence score which is linearly correlated to the segmentation accuracy on test images.
AGE2HIE: Transfer Learning from Brain Age to Predicting Neurocognitive Outcome for Infant Brain Injury
Nov 07, 2024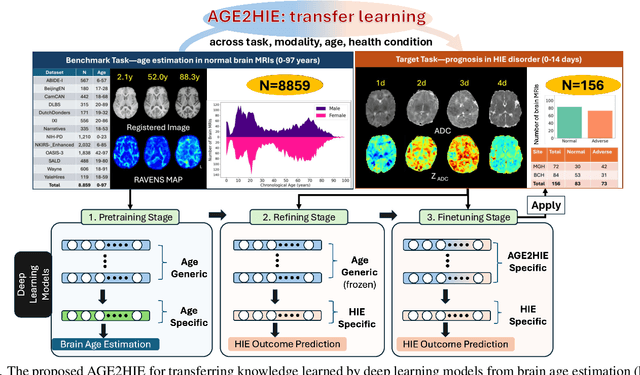
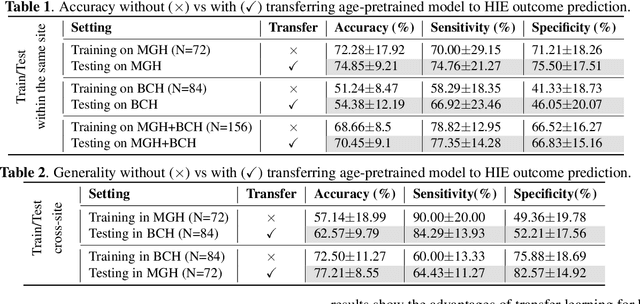
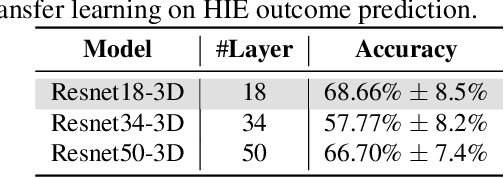
Abstract:Hypoxic-Ischemic Encephalopathy (HIE) affects 1 to 5 out of every 1,000 newborns, with 30% to 50% of cases resulting in adverse neurocognitive outcomes. However, these outcomes can only be reliably assessed as early as age 2. Therefore, early and accurate prediction of HIE-related neurocognitive outcomes using deep learning models is critical for improving clinical decision-making, guiding treatment decisions and assessing novel therapies. However, a major challenge in developing deep learning models for this purpose is the scarcity of large, annotated HIE datasets. We have assembled the first and largest public dataset, however it contains only 156 cases with 2-year neurocognitive outcome labels. In contrast, we have collected 8,859 normal brain black Magnetic Resonance Imagings (MRIs) with 0-97 years of age that are available for brain age estimation using deep learning models. In this paper, we introduce AGE2HIE to transfer knowledge learned by deep learning models from healthy controls brain MRIs to a diseased cohort, from structural to diffusion MRIs, from regression of continuous age estimation to prediction of the binary neurocognitive outcomes, and from lifespan age (0-97 years) to infant (0-2 weeks). Compared to training from scratch, transfer learning from brain age estimation significantly improves not only the prediction accuracy (3% or 2% improvement in same or multi-site), but also the model generalization across different sites (5% improvement in cross-site validation).
Foundation AI Model for Medical Image Segmentation
Nov 05, 2024Abstract:Foundation models refer to artificial intelligence (AI) models that are trained on massive amounts of data and demonstrate broad generalizability across various tasks with high accuracy. These models offer versatile, one-for-many or one-for-all solutions, eliminating the need for developing task-specific AI models. Examples of such foundation models include the Chat Generative Pre-trained Transformer (ChatGPT) and the Segment Anything Model (SAM). These models have been trained on millions to billions of samples and have shown wide-ranging and accurate applications in numerous tasks such as text processing (using ChatGPT) and natural image segmentation (using SAM). In medical image segmentation - finding target regions in medical images - there is a growing need for these one-for-many or one-for-all foundation models. Such models could obviate the need to develop thousands of task-specific AI models, which is currently standard practice in the field. They can also be adapted to tasks with datasets too small for effective training. We discuss two paths to achieve foundation models for medical image segmentation and comment on progress, challenges, and opportunities. One path is to adapt or fine-tune existing models, originally developed for natural images, for use with medical images. The second path entails building models from scratch, exclusively training on medical images.
BOston Neonatal Brain Injury Data for Hypoxic Ischemic Encephalopathy (BONBID-HIE): II. 2-year Neurocognitive Outcome and NICU Outcome
Nov 05, 2024


Abstract:Hypoxic Ischemic Encephalopathy (HIE) affects approximately 1-5/1000 newborns globally and leads to adverse neurocognitive outcomes in 30% to 50% of cases by two years of age. Despite therapeutic advances with Therapeutic Hypothermia (TH), prognosis remains challenging, highlighting the need for improved biomarkers. This paper introduces the second release of the Boston Neonatal Brain Injury Dataset for Hypoxic-Ischemic Encephalopathy (BONBID-HIE), an open-source, comprehensive MRI and clinical dataset featuring 237 patients, including NICU outcomes and 2-year neurocognitive outcomes from Massachusetts General Hospital and Boston Children's Hospital.
Accuracy of Segment-Anything Model in medical image segmentation tasks
Apr 27, 2023Abstract:The segment-anything model (SAM), was introduced as a fundamental model for segmenting images. It was trained using over 1 billion masks from 11 million natural images. The model can perform zero-shot segmentation of images by using various prompts such as masks, boxes, and points. In this report, we explored (1) the accuracy of SAM on 12 public medical image segmentation datasets which cover various organs (brain, breast, chest, lung, skin, liver, bowel, pancreas, and prostate), image modalities (2D X-ray, histology, endoscropy, and 3D MRI and CT), and health conditions (normal, lesioned). (2) if the computer vision foundational segmentation model SAM can provide promising research directions for medical image segmentation. We found that SAM without re-training on medical images does not perform as accurately as U-Net or other deep learning models trained on medical images.
U-Netmer: U-Net meets Transformer for medical image segmentation
Apr 03, 2023Abstract:The combination of the U-Net based deep learning models and Transformer is a new trend for medical image segmentation. U-Net can extract the detailed local semantic and texture information and Transformer can learn the long-rang dependencies among pixels in the input image. However, directly adapting the Transformer for segmentation has ``token-flatten" problem (flattens the local patches into 1D tokens which losses the interaction among pixels within local patches) and ``scale-sensitivity" problem (uses a fixed scale to split the input image into local patches). Compared to directly combining U-Net and Transformer, we propose a new global-local fashion combination of U-Net and Transformer, named U-Netmer, to solve the two problems. The proposed U-Netmer splits an input image into local patches. The global-context information among local patches is learnt by the self-attention mechanism in Transformer and U-Net segments each local patch instead of flattening into tokens to solve the `token-flatten" problem. The U-Netmer can segment the input image with different patch sizes with the identical structure and the same parameter. Thus, the U-Netmer can be trained with different patch sizes to solve the ``scale-sensitivity" problem. We conduct extensive experiments in 7 public datasets on 7 organs (brain, heart, breast, lung, polyp, pancreas and prostate) and 4 imaging modalities (MRI, CT, ultrasound, and endoscopy) to show that the proposed U-Netmer can be generally applied to improve accuracy of medical image segmentation. These experimental results show that U-Netmer provides state-of-the-art performance compared to baselines and other models. In addition, the discrepancy among the outputs of U-Netmer with different scales is linearly correlated to the segmentation accuracy which can be considered as a confidence score to rank test images by difficulty without ground-truth.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge