Eileen Pan
BendVLM: Test-Time Debiasing of Vision-Language Embeddings
Nov 07, 2024



Abstract:Vision-language model (VLM) embeddings have been shown to encode biases present in their training data, such as societal biases that prescribe negative characteristics to members of various racial and gender identities. VLMs are being quickly adopted for a variety of tasks ranging from few-shot classification to text-guided image generation, making debiasing VLM embeddings crucial. Debiasing approaches that fine-tune the VLM often suffer from catastrophic forgetting. On the other hand, fine-tuning-free methods typically utilize a "one-size-fits-all" approach that assumes that correlation with the spurious attribute can be explained using a single linear direction across all possible inputs. In this work, we propose Bend-VLM, a nonlinear, fine-tuning-free approach for VLM embedding debiasing that tailors the debiasing operation to each unique input. This allows for a more flexible debiasing approach. Additionally, we do not require knowledge of the set of inputs a priori to inference time, making our method more appropriate for online, open-set tasks such as retrieval and text guided image generation.
Shape-aware Segmentation of the Placenta in BOLD Fetal MRI Time Series
Dec 08, 2023


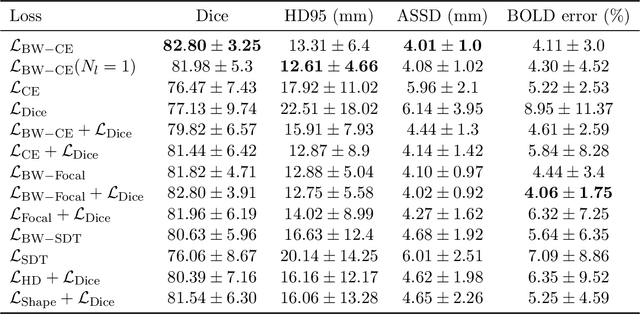
Abstract:Blood oxygen level dependent (BOLD) MRI time series with maternal hyperoxia can assess placental oxygenation and function. Measuring precise BOLD changes in the placenta requires accurate temporal placental segmentation and is confounded by fetal and maternal motion, contractions, and hyperoxia-induced intensity changes. Current BOLD placenta segmentation methods warp a manually annotated subject-specific template to the entire time series. However, as the placenta is a thin, elongated, and highly non-rigid organ subject to large deformations and obfuscated edges, existing work cannot accurately segment the placental shape, especially near boundaries. In this work, we propose a machine learning segmentation framework for placental BOLD MRI and apply it to segmenting each volume in a time series. We use a placental-boundary weighted loss formulation and perform a comprehensive evaluation across several popular segmentation objectives. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. Biomedically, our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. We further find that boundary-weighting increases placental segmentation performance by 8.3% and 6.0% Dice coefficient for the cross-entropy and signed distance transform objectives, respectively. Our code and trained model is available at https://github.com/mabulnaga/automatic-placenta-segmentation.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:017. arXiv admin note: substantial text overlap with arXiv:2208.02895
Automatic Segmentation of the Placenta in BOLD MRI Time Series
Aug 04, 2022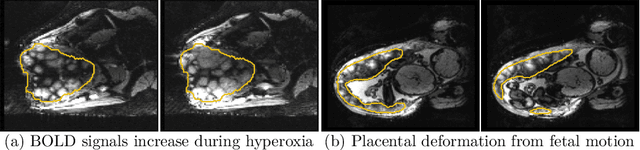
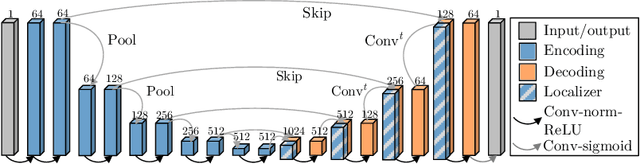

Abstract:Blood oxygen level dependent (BOLD) MRI with maternal hyperoxia can assess oxygen transport within the placenta and has emerged as a promising tool to study placental function. Measuring signal changes over time requires segmenting the placenta in each volume of the time series. Due to the large number of volumes in the BOLD time series, existing studies rely on registration to map all volumes to a manually segmented template. As the placenta can undergo large deformation due to fetal motion, maternal motion, and contractions, this approach often results in a large number of discarded volumes, where the registration approach fails. In this work, we propose a machine learning model based on a U-Net neural network architecture to automatically segment the placenta in BOLD MRI and apply it to segmenting each volume in a time series. We use a boundary-weighted loss function to accurately capture the placental shape. Our model is trained and tested on a cohort of 91 subjects containing healthy fetuses, fetuses with fetal growth restriction, and mothers with high BMI. We achieve a Dice score of 0.83+/-0.04 when matching with ground truth labels and our model performs reliably in segmenting volumes in both normoxic and hyperoxic points in the BOLD time series. Our code and trained model are available at https://github.com/mabulnaga/automatic-placenta-segmentation.
What Disease does this Patient Have? A Large-scale Open Domain Question Answering Dataset from Medical Exams
Sep 28, 2020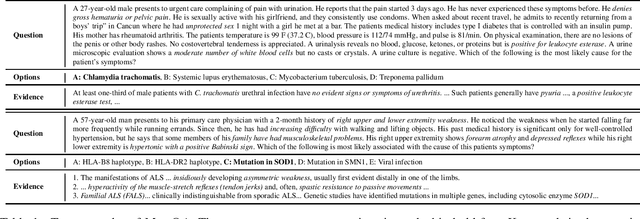
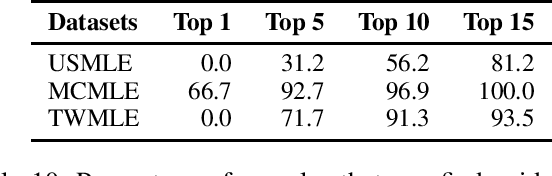

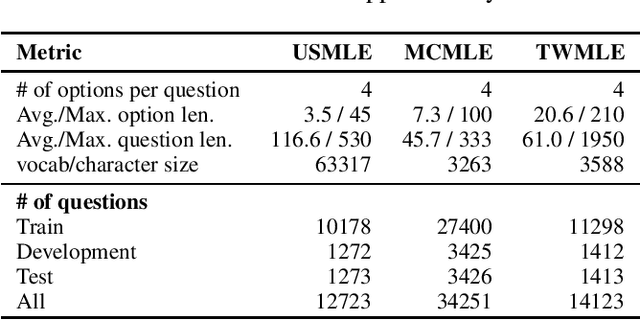
Abstract:Open domain question answering (OpenQA) tasks have been recently attracting more and more attention from the natural language processing (NLP) community. In this work, we present the first free-form multiple-choice OpenQA dataset for solving medical problems, MedQA, collected from the professional medical board exams. It covers three languages: English, simplified Chinese, and traditional Chinese, and contains 12,723, 34,251, and 14,123 questions for the three languages, respectively. We implement both rule-based and popular neural methods by sequentially combining a document retriever and a machine comprehension model. Through experiments, we find that even the current best method can only achieve 36.7\%, 42.0\%, and 70.1\% of test accuracy on the English, traditional Chinese, and simplified Chinese questions, respectively. We expect MedQA to present great challenges to existing OpenQA systems and hope that it can serve as a platform to promote much stronger OpenQA models from the NLP community in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge