Nassim Oufattole
Foundation Model of Electronic Medical Records for Adaptive Risk Estimation
Feb 10, 2025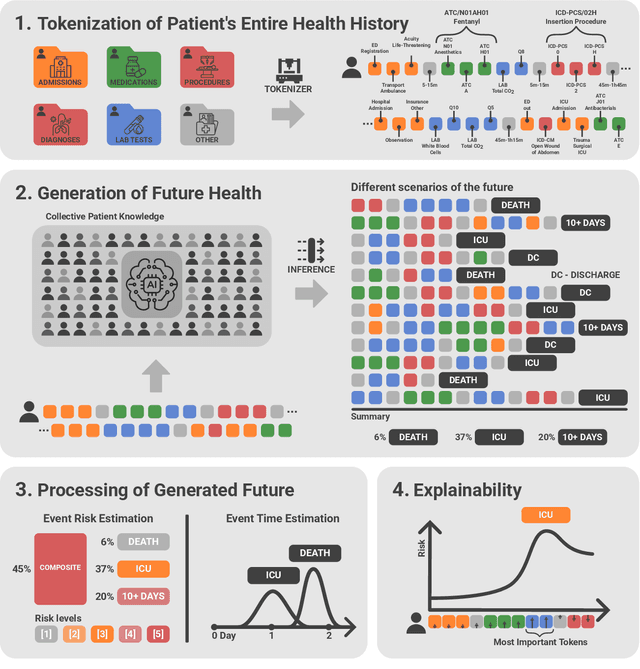
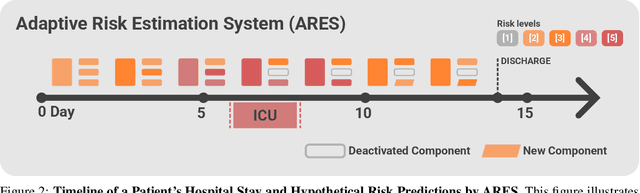
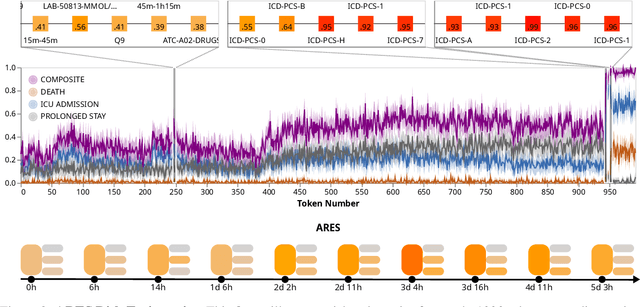
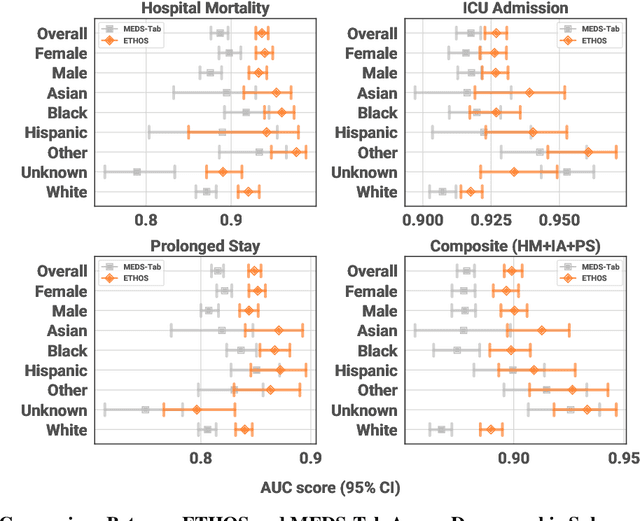
Abstract:We developed the Enhanced Transformer for Health Outcome Simulation (ETHOS), an AI model that tokenizes patient health timelines (PHTs) from EHRs. ETHOS predicts future PHTs using transformer-based architectures. The Adaptive Risk Estimation System (ARES) employs ETHOS to compute dynamic and personalized risk probabilities for clinician-defined critical events. ARES incorporates a personalized explainability module that identifies key clinical factors influencing risk estimates for individual patients. ARES was evaluated on the MIMIC-IV v2.2 dataset in emergency department (ED) settings, benchmarking its performance against traditional early warning systems and machine learning models. We processed 299,721 unique patients from MIMIC-IV into 285,622 PHTs, with 60% including hospital admissions. The dataset contained over 357 million tokens. ETHOS outperformed benchmark models in predicting hospital admissions, ICU admissions, and prolonged hospital stays, achieving superior AUC scores. ETHOS-based risk estimates demonstrated robustness across demographic subgroups with strong model reliability, confirmed via calibration curves. The personalized explainability module provides insights into patient-specific factors contributing to risk. ARES, powered by ETHOS, advances predictive healthcare AI by providing dynamic, real-time, and personalized risk estimation with patient-specific explainability to enhance clinician trust. Its adaptability and superior accuracy position it as a transformative tool for clinical decision-making, potentially improving patient outcomes and resource allocation in emergency and inpatient settings. We release the full code at github.com/ipolharvard/ethos-ares to facilitate future research.
MEDS-Tab: Automated tabularization and baseline methods for MEDS datasets
Oct 31, 2024



Abstract:Effective, reliable, and scalable development of machine learning (ML) solutions for structured electronic health record (EHR) data requires the ability to reliably generate high-quality baseline models for diverse supervised learning tasks in an efficient and performant manner. Historically, producing such baseline models has been a largely manual effort--individual researchers would need to decide on the particular featurization and tabularization processes to apply to their individual raw, longitudinal data; and then train a supervised model over those data to produce a baseline result to compare novel methods against, all for just one task and one dataset. In this work, powered by complementary advances in core data standardization through the MEDS framework, we dramatically simplify and accelerate this process of tabularizing irregularly sampled time-series data, providing researchers the ability to automatically and scalably featurize and tabularize their longitudinal EHR data across tens of thousands of individual features, hundreds of millions of clinical events, and diverse windowing horizons and aggregation strategies, all before ultimately leveraging these tabular data to automatically produce high-caliber XGBoost baselines in a highly computationally efficient manner. This system scales to dramatically larger datasets than tabularization tools currently available to the community and enables researchers with any MEDS format dataset to immediately begin producing reliable and performant baseline prediction results on various tasks, with minimal human effort required. This system will greatly enhance the reliability, reproducibility, and ease of development of powerful ML solutions for health problems across diverse datasets and clinical settings.
Event-Based Contrastive Learning for Medical Time Series
Dec 16, 2023



Abstract:In clinical practice, one often needs to identify whether a patient is at high risk of adverse outcomes after some key medical event; e.g., the short-term risk of death after an admission for heart failure. This task, however, remains challenging due to the complexity, variability, and heterogeneity of longitudinal medical data, especially for individuals suffering from chronic diseases like heart failure. In this paper, we introduce Event-Based Contrastive Learning (EBCL) - a method for learning embeddings of heterogeneous patient data that preserves temporal information before and after key index events. We demonstrate that EBCL produces models that yield better fine-tuning performance on critical downstream tasks including 30-day readmission, 1-year mortality, and 1-week length of stay relative to other representation learning methods that do not exploit temporal information surrounding key medical events.
A Knowledge Distillation Approach for Sepsis Outcome Prediction from Multivariate Clinical Time Series
Nov 16, 2023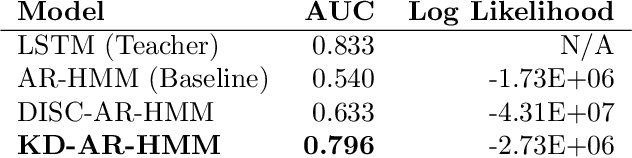
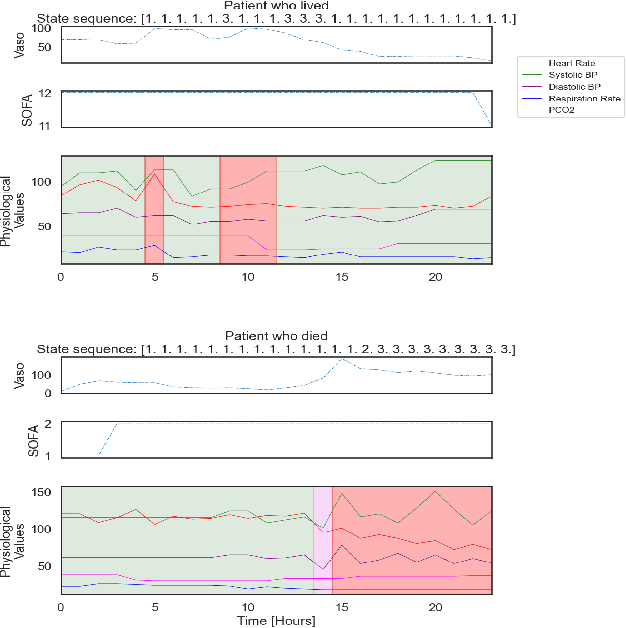
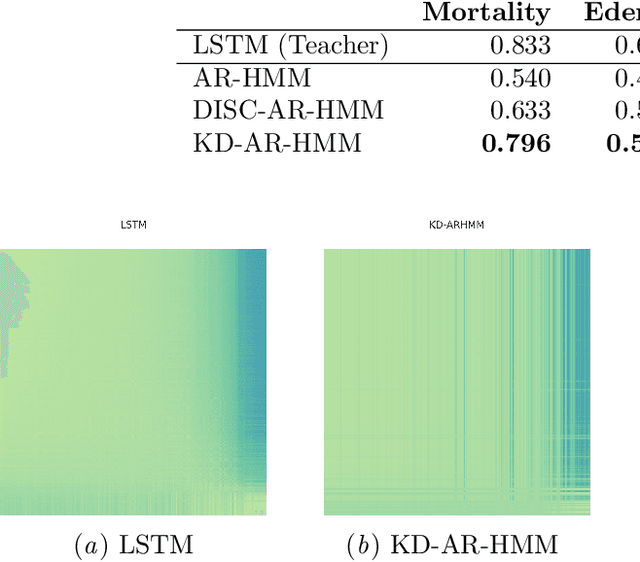

Abstract:Sepsis is a life-threatening condition triggered by an extreme infection response. Our objective is to forecast sepsis patient outcomes using their medical history and treatments, while learning interpretable state representations to assess patients' risks in developing various adverse outcomes. While neural networks excel in outcome prediction, their limited interpretability remains a key issue. In this work, we use knowledge distillation via constrained variational inference to distill the knowledge of a powerful "teacher" neural network model with high predictive power to train a "student" latent variable model to learn interpretable hidden state representations to achieve high predictive performance for sepsis outcome prediction. Using real-world data from the MIMIC-IV database, we trained an LSTM as the "teacher" model to predict mortality for sepsis patients, given information about their recent history of vital signs, lab values and treatments. For our student model, we use an autoregressive hidden Markov model (AR-HMM) to learn interpretable hidden states from patients' clinical time series, and use the posterior distribution of the learned state representations to predict various downstream outcomes, including hospital mortality, pulmonary edema, need for diuretics, dialysis, and mechanical ventilation. Our results show that our approach successfully incorporates the constraint to achieve high predictive power similar to the teacher model, while maintaining the generative performance.
What Disease does this Patient Have? A Large-scale Open Domain Question Answering Dataset from Medical Exams
Sep 28, 2020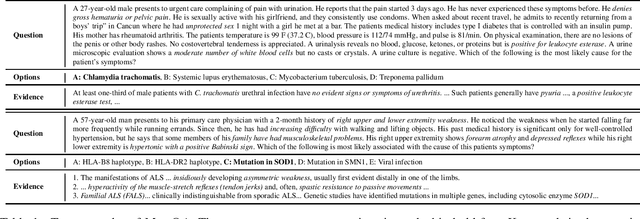
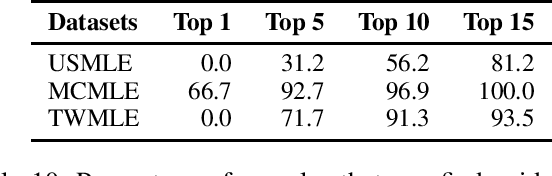

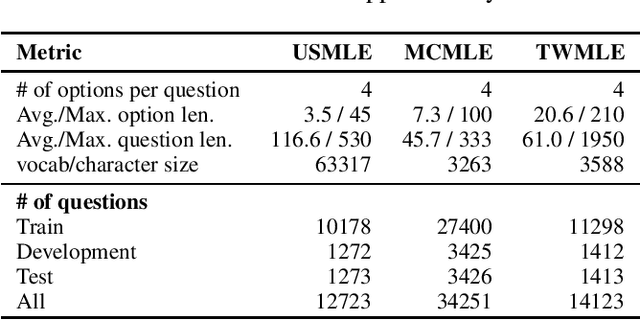
Abstract:Open domain question answering (OpenQA) tasks have been recently attracting more and more attention from the natural language processing (NLP) community. In this work, we present the first free-form multiple-choice OpenQA dataset for solving medical problems, MedQA, collected from the professional medical board exams. It covers three languages: English, simplified Chinese, and traditional Chinese, and contains 12,723, 34,251, and 14,123 questions for the three languages, respectively. We implement both rule-based and popular neural methods by sequentially combining a document retriever and a machine comprehension model. Through experiments, we find that even the current best method can only achieve 36.7\%, 42.0\%, and 70.1\% of test accuracy on the English, traditional Chinese, and simplified Chinese questions, respectively. We expect MedQA to present great challenges to existing OpenQA systems and hope that it can serve as a platform to promote much stronger OpenQA models from the NLP community in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge