Collin M. Stultz
MEDS-Tab: Automated tabularization and baseline methods for MEDS datasets
Oct 31, 2024



Abstract:Effective, reliable, and scalable development of machine learning (ML) solutions for structured electronic health record (EHR) data requires the ability to reliably generate high-quality baseline models for diverse supervised learning tasks in an efficient and performant manner. Historically, producing such baseline models has been a largely manual effort--individual researchers would need to decide on the particular featurization and tabularization processes to apply to their individual raw, longitudinal data; and then train a supervised model over those data to produce a baseline result to compare novel methods against, all for just one task and one dataset. In this work, powered by complementary advances in core data standardization through the MEDS framework, we dramatically simplify and accelerate this process of tabularizing irregularly sampled time-series data, providing researchers the ability to automatically and scalably featurize and tabularize their longitudinal EHR data across tens of thousands of individual features, hundreds of millions of clinical events, and diverse windowing horizons and aggregation strategies, all before ultimately leveraging these tabular data to automatically produce high-caliber XGBoost baselines in a highly computationally efficient manner. This system scales to dramatically larger datasets than tabularization tools currently available to the community and enables researchers with any MEDS format dataset to immediately begin producing reliable and performant baseline prediction results on various tasks, with minimal human effort required. This system will greatly enhance the reliability, reproducibility, and ease of development of powerful ML solutions for health problems across diverse datasets and clinical settings.
Deep Metric Learning for the Hemodynamics Inference with Electrocardiogram Signals
Aug 09, 2023Abstract:Heart failure is a debilitating condition that affects millions of people worldwide and has a significant impact on their quality of life and mortality rates. An objective assessment of cardiac pressures remains an important method for the diagnosis and treatment prognostication for patients with heart failure. Although cardiac catheterization is the gold standard for estimating central hemodynamic pressures, it is an invasive procedure that carries inherent risks, making it a potentially dangerous procedure for some patients. Approaches that leverage non-invasive signals - such as electrocardiogram (ECG) - have the promise to make the routine estimation of cardiac pressures feasible in both inpatient and outpatient settings. Prior models trained to estimate intracardiac pressures (e.g., mean pulmonary capillary wedge pressure (mPCWP)) in a supervised fashion have shown good discriminatory ability but have been limited to the labeled dataset from the heart failure cohort. To address this issue and build a robust representation, we apply deep metric learning (DML) and propose a novel self-supervised DML with distance-based mining that improves the performance of a model with limited labels. We use a dataset that contains over 5.4 million ECGs without concomitant central pressure labels to pre-train a self-supervised DML model which showed improved classification of elevated mPCWP compared to self-supervised contrastive baselines. Additionally, the supervised DML model that is using ECGs with access to 8,172 mPCWP labels demonstrated significantly better performance on the mPCWP regression task compared to the supervised baseline. Moreover, our data suggest that DML yields models that are performant across patient subgroups, even when some patient subgroups are under-represented in the dataset. Our code is available at https://github.com/mandiehyewon/ssldml
Sequential Multi-Dimensional Self-Supervised Learning for Clinical Time Series
Jul 20, 2023



Abstract:Self-supervised learning (SSL) for clinical time series data has received significant attention in recent literature, since these data are highly rich and provide important information about a patient's physiological state. However, most existing SSL methods for clinical time series are limited in that they are designed for unimodal time series, such as a sequence of structured features (e.g., lab values and vitals signs) or an individual high-dimensional physiological signal (e.g., an electrocardiogram). These existing methods cannot be readily extended to model time series that exhibit multimodality, with structured features and high-dimensional data being recorded at each timestep in the sequence. In this work, we address this gap and propose a new SSL method -- Sequential Multi-Dimensional SSL -- where a SSL loss is applied both at the level of the entire sequence and at the level of the individual high-dimensional data points in the sequence in order to better capture information at both scales. Our strategy is agnostic to the specific form of loss function used at each level -- it can be contrastive, as in SimCLR, or non-contrastive, as in VICReg. We evaluate our method on two real-world clinical datasets, where the time series contains sequences of (1) high-frequency electrocardiograms and (2) structured data from lab values and vitals signs. Our experimental results indicate that pre-training with our method and then fine-tuning on downstream tasks improves performance over baselines on both datasets, and in several settings, can lead to improvements across different self-supervised loss functions.
Data Augmentation for Electrocardiograms
Apr 09, 2022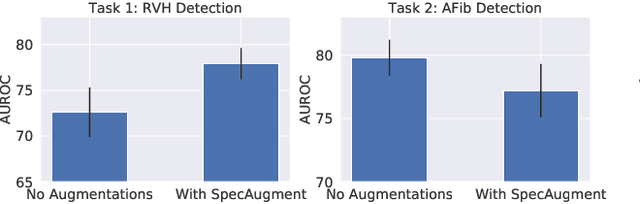
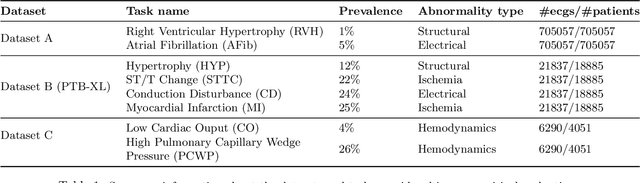
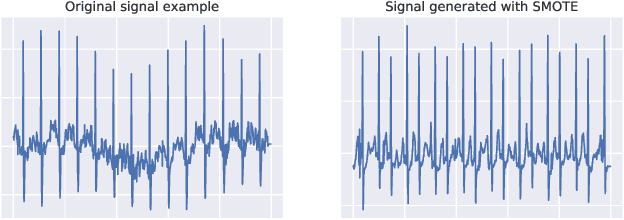
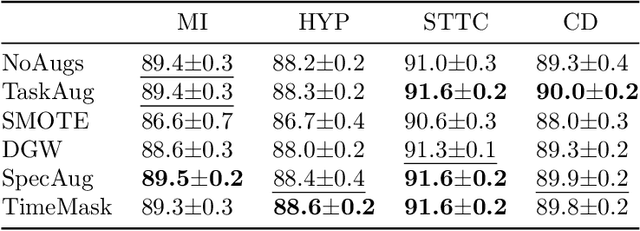
Abstract:Neural network models have demonstrated impressive performance in predicting pathologies and outcomes from the 12-lead electrocardiogram (ECG). However, these models often need to be trained with large, labelled datasets, which are not available for many predictive tasks of interest. In this work, we perform an empirical study examining whether training time data augmentation methods can be used to improve performance on such data-scarce ECG prediction problems. We investigate how data augmentation strategies impact model performance when detecting cardiac abnormalities from the ECG. Motivated by our finding that the effectiveness of existing augmentation strategies is highly task-dependent, we introduce a new method, TaskAug, which defines a flexible augmentation policy that is optimized on a per-task basis. We outline an efficient learning algorithm to do so that leverages recent work in nested optimization and implicit differentiation. In experiments, considering three datasets and eight predictive tasks, we find that TaskAug is competitive with or improves on prior work, and the learned policies shed light on what transformations are most effective for different tasks. We distill key insights from our experimental evaluation, generating a set of best practices for applying data augmentation to ECG prediction problems.
Learning to Predict with Supporting Evidence: Applications to Clinical Risk Prediction
Mar 04, 2021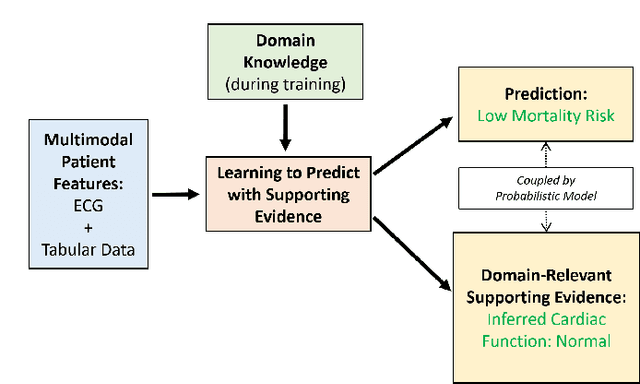
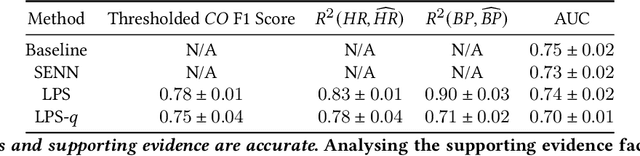
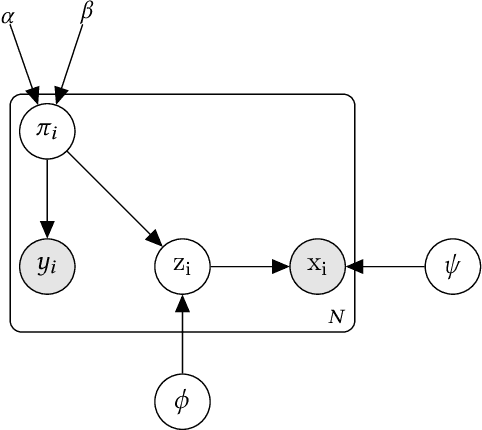
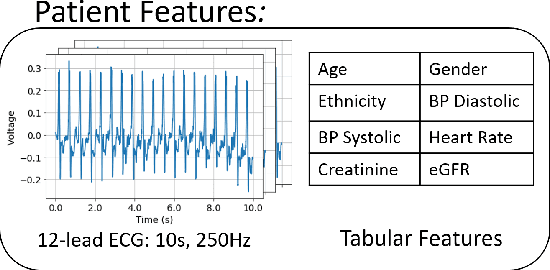
Abstract:The impact of machine learning models on healthcare will depend on the degree of trust that healthcare professionals place in the predictions made by these models. In this paper, we present a method to provide people with clinical expertise with domain-relevant evidence about why a prediction should be trusted. We first design a probabilistic model that relates meaningful latent concepts to prediction targets and observed data. Inference of latent variables in this model corresponds to both making a prediction and providing supporting evidence for that prediction. We present a two-step process to efficiently approximate inference: (i) estimating model parameters using variational learning, and (ii) approximating maximum a posteriori estimation of latent variables in the model using a neural network, trained with an objective derived from the probabilistic model. We demonstrate the method on the task of predicting mortality risk for patients with cardiovascular disease. Specifically, using electrocardiogram and tabular data as input, we show that our approach provides appropriate domain-relevant supporting evidence for accurate predictions.
Transferring Knowledge from Text to Predict Disease Onset
Aug 06, 2016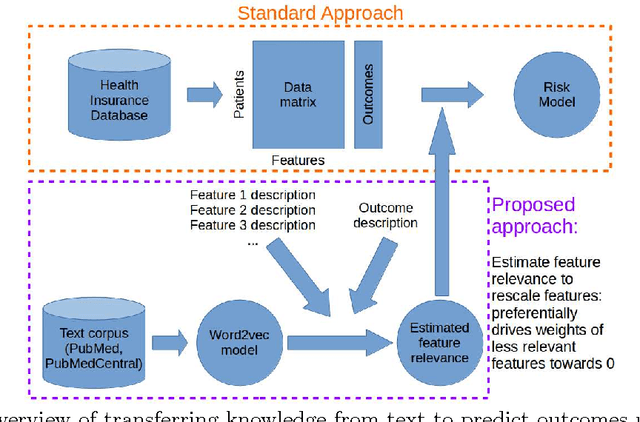
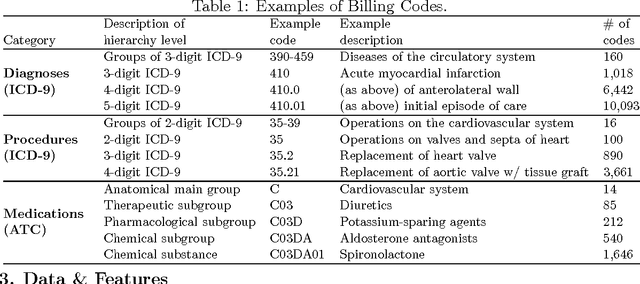
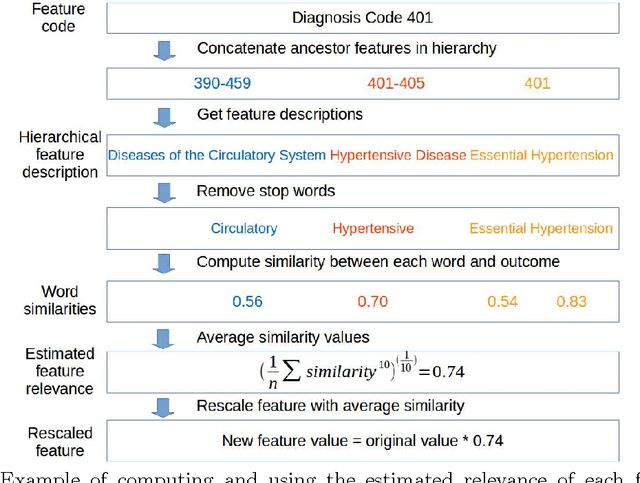
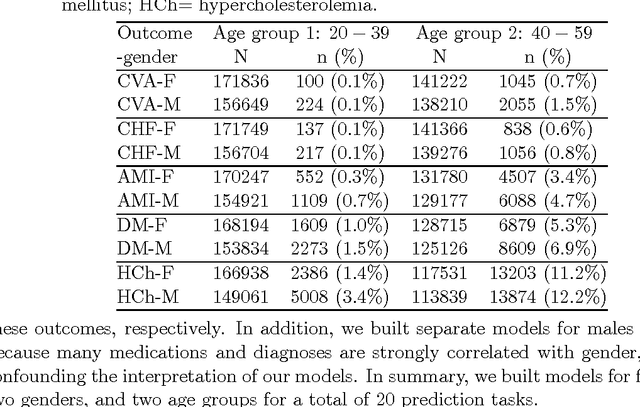
Abstract:In many domains such as medicine, training data is in short supply. In such cases, external knowledge is often helpful in building predictive models. We propose a novel method to incorporate publicly available domain expertise to build accurate models. Specifically, we use word2vec models trained on a domain-specific corpus to estimate the relevance of each feature's text description to the prediction problem. We use these relevance estimates to rescale the features, causing more important features to experience weaker regularization. We apply our method to predict the onset of five chronic diseases in the next five years in two genders and two age groups. Our rescaling approach improves the accuracy of the model, particularly when there are few positive examples. Furthermore, our method selects 60% fewer features, easing interpretation by physicians. Our method is applicable to other domains where feature and outcome descriptions are available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge