Aniruddh Raghu
A Unification of Discrete, Gaussian, and Simplicial Diffusion
Dec 17, 2025Abstract:To model discrete sequences such as DNA, proteins, and language using diffusion, practitioners must choose between three major methods: diffusion in discrete space, Gaussian diffusion in Euclidean space, or diffusion on the simplex. Despite their shared goal, these models have disparate algorithms, theoretical structures, and tradeoffs: discrete diffusion has the most natural domain, Gaussian diffusion has more mature algorithms, and diffusion on the simplex in principle combines the strengths of the other two but in practice suffers from a numerically unstable stochastic processes. Ideally we could see each of these models as instances of the same underlying framework, and enable practitioners to switch between models for downstream applications. However previous theories have only considered connections in special cases. Here we build a theory unifying all three methods of discrete diffusion as different parameterizations of the same underlying process: the Wright-Fisher population genetics model. In particular, we find simplicial and Gaussian diffusion as two large-population limits. Our theory formally connects the likelihoods and hyperparameters of these models and leverages decades of mathematical genetics literature to unlock stable simplicial diffusion. Finally, we relieve the practitioner of balancing model trade-offs by demonstrating it is possible to train a single model that can perform diffusion in any of these three domains at test time. Our experiments show that Wright-Fisher simplicial diffusion is more stable and outperforms previous simplicial diffusion models on conditional DNA generation. We also show that we can train models on multiple domains at once that are competitive with models trained on any individual domain.
Bayesian Optimization of Antibodies Informed by a Generative Model of Evolving Sequences
Dec 10, 2024Abstract:To build effective therapeutics, biologists iteratively mutate antibody sequences to improve binding and stability. Proposed mutations can be informed by previous measurements or by learning from large antibody databases to predict only typical antibodies. Unfortunately, the space of typical antibodies is enormous to search, and experiments often fail to find suitable antibodies on a budget. We introduce Clone-informed Bayesian Optimization (CloneBO), a Bayesian optimization procedure that efficiently optimizes antibodies in the lab by teaching a generative model how our immune system optimizes antibodies. Our immune system makes antibodies by iteratively evolving specific portions of their sequences to bind their target strongly and stably, resulting in a set of related, evolving sequences known as a clonal family. We train a large language model, CloneLM, on hundreds of thousands of clonal families and use it to design sequences with mutations that are most likely to optimize an antibody within the human immune system. We propose to guide our designs to fit previous measurements with a twisted sequential Monte Carlo procedure. We show that CloneBO optimizes antibodies substantially more efficiently than previous methods in realistic in silico experiments and designs stronger and more stable binders in in vitro wet lab experiments.
Generative Humanization for Therapeutic Antibodies
Dec 09, 2024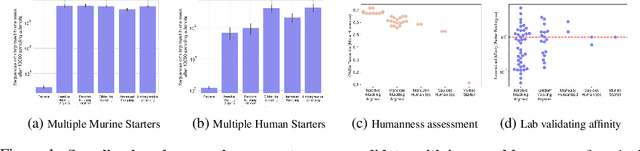
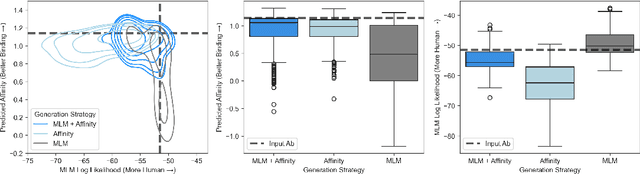
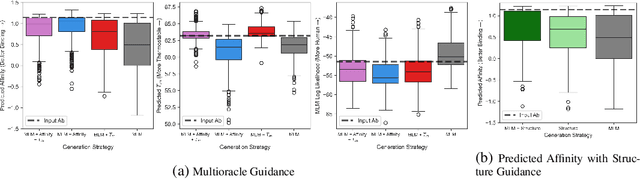
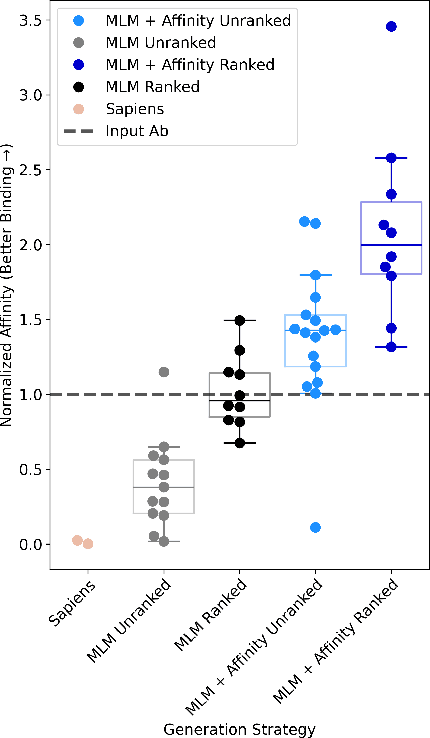
Abstract:Antibody therapies have been employed to address some of today's most challenging diseases, but must meet many criteria during drug development before reaching a patient. Humanization is a sequence optimization strategy that addresses one critical risk called immunogenicity - a patient's immune response to the drug - by making an antibody more "human-like" in the absence of a predictive lab-based test for immunogenicity. However, existing humanization strategies generally yield very few humanized candidates, which may have degraded biophysical properties or decreased drug efficacy. Here, we re-frame humanization as a conditional generative modeling task, where humanizing mutations are sampled from a language model trained on human antibody data. We describe a sampling process that incorporates models of therapeutic attributes, such as antigen binding affinity, to obtain candidate sequences that have both reduced immunogenicity risk and maintained or improved therapeutic properties, allowing this algorithm to be readily embedded into an iterative antibody optimization campaign. We demonstrate in silico and in lab validation that in real therapeutic programs our generative humanization method produces diverse sets of antibodies that are both (1) highly-human and (2) have favorable therapeutic properties, such as improved binding to target antigens.
Sequential Multi-Dimensional Self-Supervised Learning for Clinical Time Series
Jul 20, 2023



Abstract:Self-supervised learning (SSL) for clinical time series data has received significant attention in recent literature, since these data are highly rich and provide important information about a patient's physiological state. However, most existing SSL methods for clinical time series are limited in that they are designed for unimodal time series, such as a sequence of structured features (e.g., lab values and vitals signs) or an individual high-dimensional physiological signal (e.g., an electrocardiogram). These existing methods cannot be readily extended to model time series that exhibit multimodality, with structured features and high-dimensional data being recorded at each timestep in the sequence. In this work, we address this gap and propose a new SSL method -- Sequential Multi-Dimensional SSL -- where a SSL loss is applied both at the level of the entire sequence and at the level of the individual high-dimensional data points in the sequence in order to better capture information at both scales. Our strategy is agnostic to the specific form of loss function used at each level -- it can be contrastive, as in SimCLR, or non-contrastive, as in VICReg. We evaluate our method on two real-world clinical datasets, where the time series contains sequences of (1) high-frequency electrocardiograms and (2) structured data from lab values and vitals signs. Our experimental results indicate that pre-training with our method and then fine-tuning on downstream tasks improves performance over baselines on both datasets, and in several settings, can lead to improvements across different self-supervised loss functions.
Data Augmentation for Electrocardiograms
Apr 09, 2022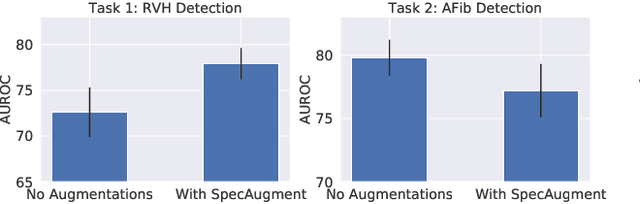
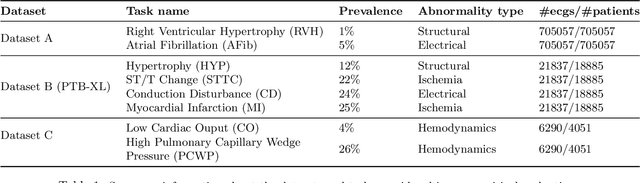
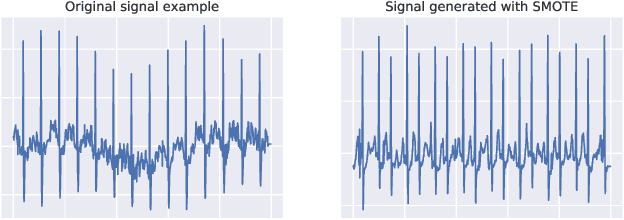
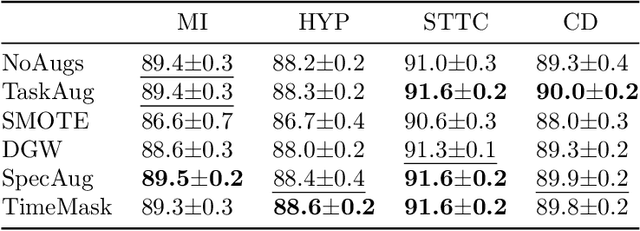
Abstract:Neural network models have demonstrated impressive performance in predicting pathologies and outcomes from the 12-lead electrocardiogram (ECG). However, these models often need to be trained with large, labelled datasets, which are not available for many predictive tasks of interest. In this work, we perform an empirical study examining whether training time data augmentation methods can be used to improve performance on such data-scarce ECG prediction problems. We investigate how data augmentation strategies impact model performance when detecting cardiac abnormalities from the ECG. Motivated by our finding that the effectiveness of existing augmentation strategies is highly task-dependent, we introduce a new method, TaskAug, which defines a flexible augmentation policy that is optimized on a per-task basis. We outline an efficient learning algorithm to do so that leverages recent work in nested optimization and implicit differentiation. In experiments, considering three datasets and eight predictive tasks, we find that TaskAug is competitive with or improves on prior work, and the learned policies shed light on what transformations are most effective for different tasks. We distill key insights from our experimental evaluation, generating a set of best practices for applying data augmentation to ECG prediction problems.
Meta-Learning to Improve Pre-Training
Nov 02, 2021

Abstract:Pre-training (PT) followed by fine-tuning (FT) is an effective method for training neural networks, and has led to significant performance improvements in many domains. PT can incorporate various design choices such as task and data reweighting strategies, augmentation policies, and noise models, all of which can significantly impact the quality of representations learned. The hyperparameters introduced by these strategies therefore must be tuned appropriately. However, setting the values of these hyperparameters is challenging. Most existing methods either struggle to scale to high dimensions, are too slow and memory-intensive, or cannot be directly applied to the two-stage PT and FT learning process. In this work, we propose an efficient, gradient-based algorithm to meta-learn PT hyperparameters. We formalize the PT hyperparameter optimization problem and propose a novel method to obtain PT hyperparameter gradients by combining implicit differentiation and backpropagation through unrolled optimization. We demonstrate that our method improves predictive performance on two real-world domains. First, we optimize high-dimensional task weighting hyperparameters for multitask pre-training on protein-protein interaction graphs and improve AUROC by up to 3.9%. Second, we optimize a data augmentation neural network for self-supervised PT with SimCLR on electrocardiography data and improve AUROC by up to 1.9%.
Learning to Predict with Supporting Evidence: Applications to Clinical Risk Prediction
Mar 04, 2021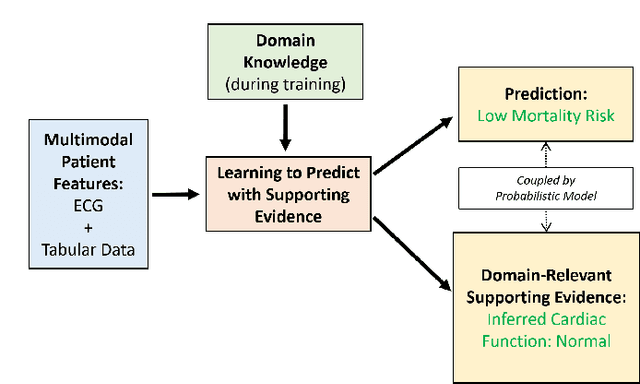
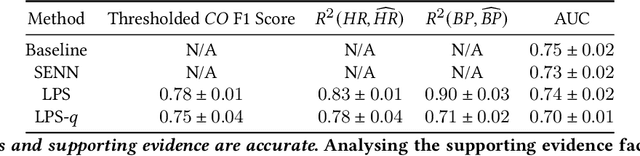
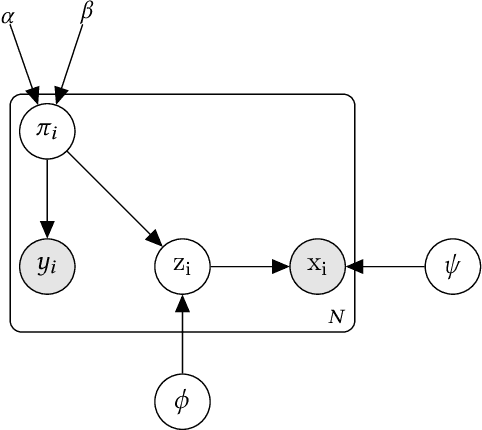
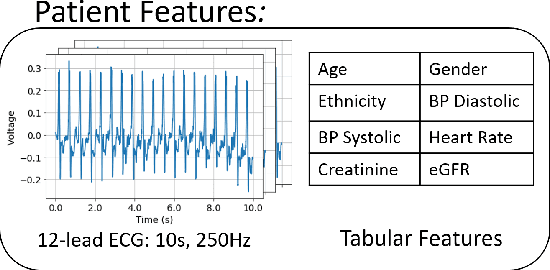
Abstract:The impact of machine learning models on healthcare will depend on the degree of trust that healthcare professionals place in the predictions made by these models. In this paper, we present a method to provide people with clinical expertise with domain-relevant evidence about why a prediction should be trusted. We first design a probabilistic model that relates meaningful latent concepts to prediction targets and observed data. Inference of latent variables in this model corresponds to both making a prediction and providing supporting evidence for that prediction. We present a two-step process to efficiently approximate inference: (i) estimating model parameters using variational learning, and (ii) approximating maximum a posteriori estimation of latent variables in the model using a neural network, trained with an objective derived from the probabilistic model. We demonstrate the method on the task of predicting mortality risk for patients with cardiovascular disease. Specifically, using electrocardiogram and tabular data as input, we show that our approach provides appropriate domain-relevant supporting evidence for accurate predictions.
Teaching with Commentaries
Nov 05, 2020



Abstract:Effective training of deep neural networks can be challenging, and there remain many open questions on how to best learn these models. Recently developed methods to improve neural network training examine teaching: providing learned information during the training process to improve downstream model performance. In this paper, we take steps towards extending the scope of teaching. We propose a flexible teaching framework using commentaries, meta-learned information helpful for training on a particular task or dataset. We present an efficient and scalable gradient-based method to learn commentaries, leveraging recent work on implicit differentiation. We explore diverse applications of commentaries, from learning weights for individual training examples, to parameterizing label-dependent data augmentation policies, to representing attention masks that highlight salient image regions. In these settings, we find that commentaries can improve training speed and/or performance and also provide fundamental insights about the dataset and training process.
Rapid Learning or Feature Reuse? Towards Understanding the Effectiveness of MAML
Sep 19, 2019



Abstract:An important research direction in machine learning has centered around developing meta-learning algorithms to tackle few-shot learning. An especially successful algorithm has been Model Agnostic Meta-Learning (MAML), a method that consists of two optimization loops, with the outer loop finding a meta-initialization, from which the inner loop can efficiently learn new tasks. Despite MAML's popularity, a fundamental open question remains -- is the effectiveness of MAML due to the meta-initialization being primed for rapid learning (large, efficient changes in the representations) or due to feature reuse, with the meta initialization already containing high quality features? We investigate this question, via ablation studies and analysis of the latent representations, finding that feature reuse is the dominant factor. This leads to the ANIL (Almost No Inner Loop) algorithm, a simplification of MAML where we remove the inner loop for all but the (task-specific) head of a MAML-trained network. ANIL matches MAML's performance on benchmark few-shot image classification and RL and offers computational improvements over MAML. We further study the precise contributions of the head and body of the network, showing that performance on the test tasks is entirely determined by the quality of the learned features, and we can remove even the head of the network (the NIL algorithm). We conclude with a discussion of the rapid learning vs feature reuse question for meta-learning algorithms more broadly.
Model-Based Reinforcement Learning for Sepsis Treatment
Nov 23, 2018
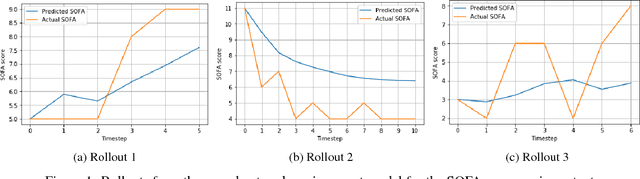

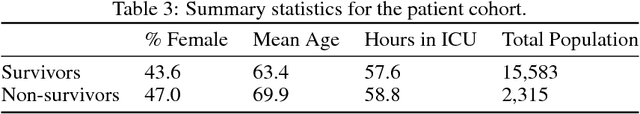
Abstract:Sepsis is a dangerous condition that is a leading cause of patient mortality. Treating sepsis is highly challenging, because individual patients respond very differently to medical interventions and there is no universally agreed-upon treatment for sepsis. In this work, we explore the use of continuous state-space model-based reinforcement learning (RL) to discover high-quality treatment policies for sepsis patients. Our quantitative evaluation reveals that by blending the treatment strategy discovered with RL with what clinicians follow, we can obtain improved policies, potentially allowing for better medical treatment for sepsis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge