Nate Gruver
Why Masking Diffusion Works: Condition on the Jump Schedule for Improved Discrete Diffusion
Jun 10, 2025Abstract:Discrete diffusion models, like continuous diffusion models, generate high-quality samples by gradually undoing noise applied to datapoints with a Markov process. Gradual generation in theory comes with many conceptual benefits; for example, inductive biases can be incorporated into the noising Markov process, and access to improved sampling algorithms. In practice, however, the consistently best performing discrete diffusion model is, surprisingly, masking diffusion, which does not denoise gradually. Here we explain the superior performance of masking diffusion by noting that it makes use of a fundamental difference between continuous and discrete Markov processes: discrete Markov processes evolve by discontinuous jumps at a fixed rate and, unlike other discrete diffusion models, masking diffusion builds in the known distribution of jump times and only learns where to jump to. We show that we can similarly bake in the known distribution of jump times into any discrete diffusion model. The resulting models - schedule-conditioned discrete diffusion (SCUD) - generalize classical discrete diffusion and masking diffusion. By applying SCUD to models with noising processes that incorporate inductive biases on images, text, and protein data, we build models that outperform masking.
Bayesian Optimization of Antibodies Informed by a Generative Model of Evolving Sequences
Dec 10, 2024Abstract:To build effective therapeutics, biologists iteratively mutate antibody sequences to improve binding and stability. Proposed mutations can be informed by previous measurements or by learning from large antibody databases to predict only typical antibodies. Unfortunately, the space of typical antibodies is enormous to search, and experiments often fail to find suitable antibodies on a budget. We introduce Clone-informed Bayesian Optimization (CloneBO), a Bayesian optimization procedure that efficiently optimizes antibodies in the lab by teaching a generative model how our immune system optimizes antibodies. Our immune system makes antibodies by iteratively evolving specific portions of their sequences to bind their target strongly and stably, resulting in a set of related, evolving sequences known as a clonal family. We train a large language model, CloneLM, on hundreds of thousands of clonal families and use it to design sequences with mutations that are most likely to optimize an antibody within the human immune system. We propose to guide our designs to fit previous measurements with a twisted sequential Monte Carlo procedure. We show that CloneBO optimizes antibodies substantially more efficiently than previous methods in realistic in silico experiments and designs stronger and more stable binders in in vitro wet lab experiments.
Large Language Models Must Be Taught to Know What They Don't Know
Jun 12, 2024



Abstract:When using large language models (LLMs) in high-stakes applications, we need to know when we can trust their predictions. Some works argue that prompting high-performance LLMs is sufficient to produce calibrated uncertainties, while others introduce sampling methods that can be prohibitively expensive. In this work, we first argue that prompting on its own is insufficient to achieve good calibration and then show that fine-tuning on a small dataset of correct and incorrect answers can create an uncertainty estimate with good generalization and small computational overhead. We show that a thousand graded examples are sufficient to outperform baseline methods and that training through the features of a model is necessary for good performance and tractable for large open-source models when using LoRA. We also investigate the mechanisms that enable reliable LLM uncertainty estimation, finding that many models can be used as general-purpose uncertainty estimators, applicable not just to their own uncertainties but also the uncertainty of other models. Lastly, we show that uncertainty estimates inform human use of LLMs in human-AI collaborative settings through a user study.
Fine-Tuned Language Models Generate Stable Inorganic Materials as Text
Feb 06, 2024Abstract:We propose fine-tuning large language models for generation of stable materials. While unorthodox, fine-tuning large language models on text-encoded atomistic data is simple to implement yet reliable, with around 90% of sampled structures obeying physical constraints on atom positions and charges. Using energy above hull calculations from both learned ML potentials and gold-standard DFT calculations, we show that our strongest model (fine-tuned LLaMA-2 70B) can generate materials predicted to be metastable at about twice the rate (49% vs 28%) of CDVAE, a competing diffusion model. Because of text prompting's inherent flexibility, our models can simultaneously be used for unconditional generation of stable material, infilling of partial structures and text-conditional generation. Finally, we show that language models' ability to capture key symmetries of crystal structures improves with model scale, suggesting that the biases of pretrained LLMs are surprisingly well-suited for atomistic data.
Large Language Models Are Zero-Shot Time Series Forecasters
Oct 11, 2023Abstract:By encoding time series as a string of numerical digits, we can frame time series forecasting as next-token prediction in text. Developing this approach, we find that large language models (LLMs) such as GPT-3 and LLaMA-2 can surprisingly zero-shot extrapolate time series at a level comparable to or exceeding the performance of purpose-built time series models trained on the downstream tasks. To facilitate this performance, we propose procedures for effectively tokenizing time series data and converting discrete distributions over tokens into highly flexible densities over continuous values. We argue the success of LLMs for time series stems from their ability to naturally represent multimodal distributions, in conjunction with biases for simplicity, and repetition, which align with the salient features in many time series, such as repeated seasonal trends. We also show how LLMs can naturally handle missing data without imputation through non-numerical text, accommodate textual side information, and answer questions to help explain predictions. While we find that increasing model size generally improves performance on time series, we show GPT-4 can perform worse than GPT-3 because of how it tokenizes numbers, and poor uncertainty calibration, which is likely the result of alignment interventions such as RLHF.
Protein Design with Guided Discrete Diffusion
May 31, 2023Abstract:A popular approach to protein design is to combine a generative model with a discriminative model for conditional sampling. The generative model samples plausible sequences while the discriminative model guides a search for sequences with high fitness. Given its broad success in conditional sampling, classifier-guided diffusion modeling is a promising foundation for protein design, leading many to develop guided diffusion models for structure with inverse folding to recover sequences. In this work, we propose diffusioN Optimized Sampling (NOS), a guidance method for discrete diffusion models that follows gradients in the hidden states of the denoising network. NOS makes it possible to perform design directly in sequence space, circumventing significant limitations of structure-based methods, including scarce data and challenging inverse design. Moreover, we use NOS to generalize LaMBO, a Bayesian optimization procedure for sequence design that facilitates multiple objectives and edit-based constraints. The resulting method, LaMBO-2, enables discrete diffusions and stronger performance with limited edits through a novel application of saliency maps. We apply LaMBO-2 to a real-world protein design task, optimizing antibodies for higher expression yield and binding affinity to a therapeutic target under locality and liability constraints, with 97% expression rate and 25% binding rate in exploratory in vitro experiments.
On Feature Learning in the Presence of Spurious Correlations
Oct 20, 2022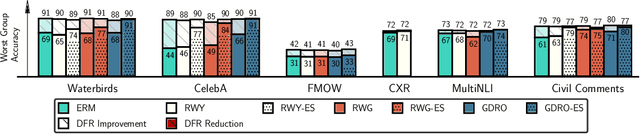
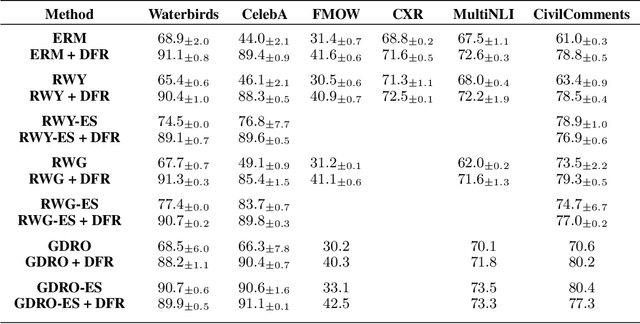
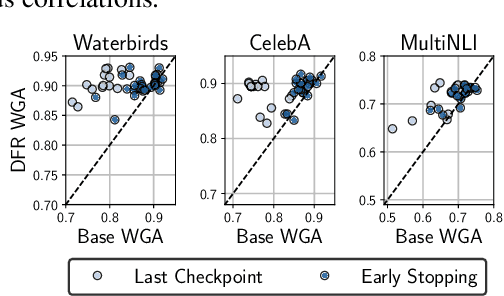
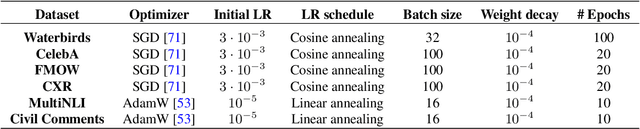
Abstract:Deep classifiers are known to rely on spurious features $\unicode{x2013}$ patterns which are correlated with the target on the training data but not inherently relevant to the learning problem, such as the image backgrounds when classifying the foregrounds. In this paper we evaluate the amount of information about the core (non-spurious) features that can be decoded from the representations learned by standard empirical risk minimization (ERM) and specialized group robustness training. Following recent work on Deep Feature Reweighting (DFR), we evaluate the feature representations by re-training the last layer of the model on a held-out set where the spurious correlation is broken. On multiple vision and NLP problems, we show that the features learned by simple ERM are highly competitive with the features learned by specialized group robustness methods targeted at reducing the effect of spurious correlations. Moreover, we show that the quality of learned feature representations is greatly affected by the design decisions beyond the training method, such as the model architecture and pre-training strategy. On the other hand, we find that strong regularization is not necessary for learning high quality feature representations. Finally, using insights from our analysis, we significantly improve upon the best results reported in the literature on the popular Waterbirds, CelebA hair color prediction and WILDS-FMOW problems, achieving 97%, 92% and 50% worst-group accuracies, respectively.
The Lie Derivative for Measuring Learned Equivariance
Oct 06, 2022



Abstract:Equivariance guarantees that a model's predictions capture key symmetries in data. When an image is translated or rotated, an equivariant model's representation of that image will translate or rotate accordingly. The success of convolutional neural networks has historically been tied to translation equivariance directly encoded in their architecture. The rising success of vision transformers, which have no explicit architectural bias towards equivariance, challenges this narrative and suggests that augmentations and training data might also play a significant role in their performance. In order to better understand the role of equivariance in recent vision models, we introduce the Lie derivative, a method for measuring equivariance with strong mathematical foundations and minimal hyperparameters. Using the Lie derivative, we study the equivariance properties of hundreds of pretrained models, spanning CNNs, transformers, and Mixer architectures. The scale of our analysis allows us to separate the impact of architecture from other factors like model size or training method. Surprisingly, we find that many violations of equivariance can be linked to spatial aliasing in ubiquitous network layers, such as pointwise non-linearities, and that as models get larger and more accurate they tend to display more equivariance, regardless of architecture. For example, transformers can be more equivariant than convolutional neural networks after training.
Accelerating Bayesian Optimization for Biological Sequence Design with Denoising Autoencoders
Mar 23, 2022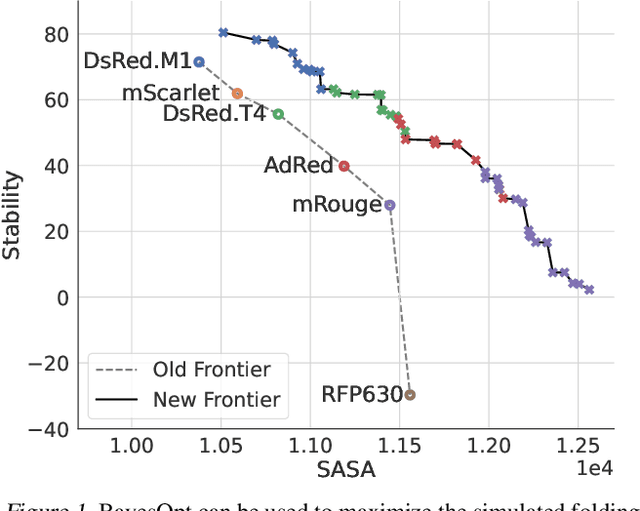
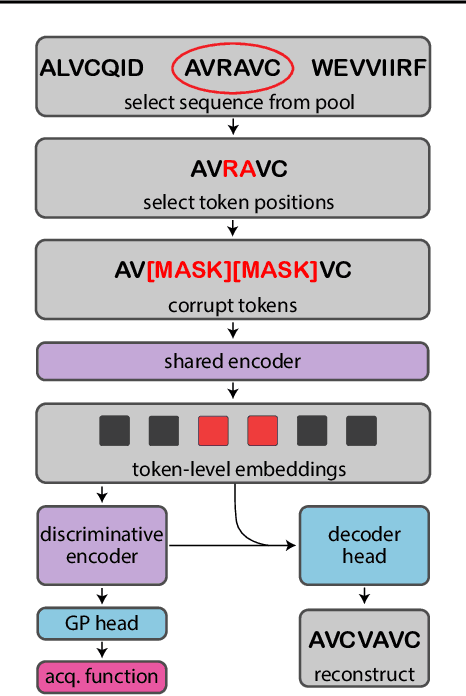
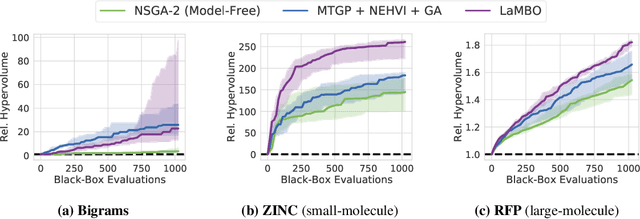
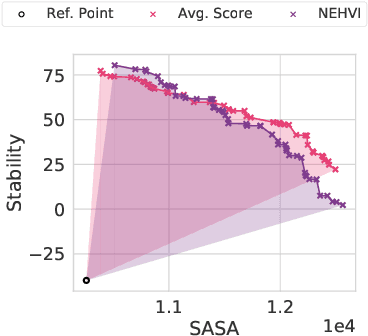
Abstract:Bayesian optimization is a gold standard for query-efficient continuous optimization. However, its adoption for drug and antibody sequence design has been hindered by the discrete, high-dimensional nature of the decision variables. We develop a new approach (LaMBO) which jointly trains a denoising autoencoder with a discriminative multi-task Gaussian process head, enabling gradient-based optimization of multi-objective acquisition functions in the latent space of the autoencoder. These acquisition functions allow LaMBO to balance the explore-exploit trade-off over multiple design rounds, and to balance objective tradeoffs by optimizing sequences at many different points on the Pareto frontier. We evaluate LaMBO on a small-molecule task based on the ZINC dataset and introduce a new large-molecule task targeting fluorescent proteins. In our experiments, LaMBO outperforms genetic optimizers and does not require a large pretraining corpus, demonstrating that Bayesian optimization is practical and effective for biological sequence design.
Deconstructing the Inductive Biases of Hamiltonian Neural Networks
Feb 12, 2022



Abstract:Physics-inspired neural networks (NNs), such as Hamiltonian or Lagrangian NNs, dramatically outperform other learned dynamics models by leveraging strong inductive biases. These models, however, are challenging to apply to many real world systems, such as those that don't conserve energy or contain contacts, a common setting for robotics and reinforcement learning. In this paper, we examine the inductive biases that make physics-inspired models successful in practice. We show that, contrary to conventional wisdom, the improved generalization of HNNs is the result of modeling acceleration directly and avoiding artificial complexity from the coordinate system, rather than symplectic structure or energy conservation. We show that by relaxing the inductive biases of these models, we can match or exceed performance on energy-conserving systems while dramatically improving performance on practical, non-conservative systems. We extend this approach to constructing transition models for common Mujoco environments, showing that our model can appropriately balance inductive biases with the flexibility required for model-based control.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge