Jaejin Cho
RestoreGrad: Signal Restoration Using Conditional Denoising Diffusion Models with Jointly Learned Prior
Feb 19, 2025Abstract:Denoising diffusion probabilistic models (DDPMs) can be utilized for recovering a clean signal from its degraded observation(s) by conditioning the model on the degraded signal. The degraded signals are themselves contaminated versions of the clean signals; due to this correlation, they may encompass certain useful information about the target clean data distribution. However, existing adoption of the standard Gaussian as the prior distribution in turn discards such information, resulting in sub-optimal performance. In this paper, we propose to improve conditional DDPMs for signal restoration by leveraging a more informative prior that is jointly learned with the diffusion model. The proposed framework, called RestoreGrad, seamlessly integrates DDPMs into the variational autoencoder framework and exploits the correlation between the degraded and clean signals to encode a better diffusion prior. On speech and image restoration tasks, we show that RestoreGrad demonstrates faster convergence (5-10 times fewer training steps) to achieve better quality of restored signals over existing DDPM baselines, and improved robustness to using fewer sampling steps in inference time (2-2.5 times fewer), advocating the advantages of leveraging jointly learned prior for efficiency improvements in the diffusion process.
PRIME: Phase Reversed Interleaved Multi-Echo acquisition enables highly accelerated distortion-free diffusion MRI
Sep 11, 2024


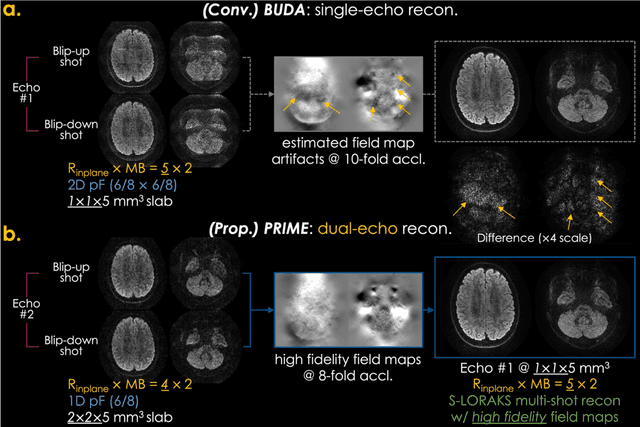
Abstract:Purpose: To develop and evaluate a new pulse sequence for highly accelerated distortion-free diffusion MRI (dMRI) by inserting an additional echo without prolonging TR, when generalized slice dithered enhanced resolution (gSlider) radiofrequency encoding is used for volumetric acquisition. Methods: A phase-reversed interleaved multi-echo acquisition (PRIME) was developed for rapid, high-resolution, and distortion-free dMRI, which includes two echoes where the first echo is for target diffusion-weighted imaging (DWI) acquisition with high-resolution and the second echo is acquired with either 1) lower-resolution for high-fidelity field map estimation, or 2) matching resolution to enable efficient diffusion relaxometry acquisitions. The sequence was evaluated on in vivo data acquired from healthy volunteers on clinical and Connectome 2.0 scanners. Results: In vivo experiments demonstrated that 1) high in-plane acceleration (Rin-plane of 5-fold with 2D partial Fourier) was achieved using the high-fidelity field maps estimated from the second echo, which was made at a lower resolution/acceleration to increase its SNR while matching the effective echo spacing of the first readout, 2) high-resolution diffusion relaxometry parameters were estimated from dual-echo PRIME data using a white matter model of multi-TE spherical mean technique (MTE-SMT), and 3) high-fidelity mesoscale DWI at 550 um isotropic resolution could be obtained in vivo by capitalizing on the high-performance gradients of the Connectome 2.0 scanner. Conclusion: The proposed PRIME sequence enabled highly accelerated, high-resolution, and distortion-free dMRI using an additional echo without prolonging scan time when gSlider encoding is utilized.
NLCG-Net: A Model-Based Zero-Shot Learning Framework for Undersampled Quantitative MRI Reconstruction
Jan 22, 2024Abstract:Typical quantitative MRI (qMRI) methods estimate parameter maps after image reconstructing, which is prone to biases and error propagation. We propose a Nonlinear Conjugate Gradient (NLCG) optimizer for model-based T2/T1 estimation, which incorporates U-Net regularization trained in a scan-specific manner. This end-to-end method directly estimates qMRI maps from undersampled k-space data using mono-exponential signal modeling with zero-shot scan-specific neural network regularization to enable high fidelity T1 and T2 mapping. T2 and T1 mapping results demonstrate the ability of the proposed NLCG-Net to improve estimation quality compared to subspace reconstruction at high accelerations.
CWCL: Cross-Modal Transfer with Continuously Weighted Contrastive Loss
Sep 26, 2023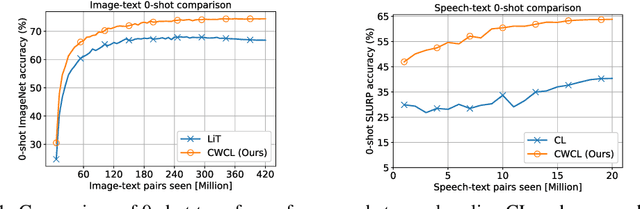
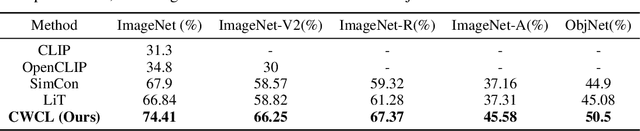
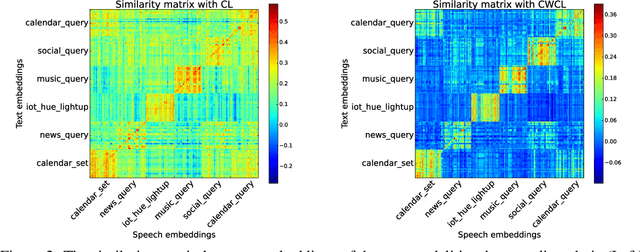

Abstract:This paper considers contrastive training for cross-modal 0-shot transfer wherein a pre-trained model in one modality is used for representation learning in another domain using pairwise data. The learnt models in the latter domain can then be used for a diverse set of tasks in a zero-shot way, similar to ``Contrastive Language-Image Pre-training (CLIP)'' and ``Locked-image Tuning (LiT)'' that have recently gained considerable attention. Most existing works for cross-modal representation alignment (including CLIP and LiT) use the standard contrastive training objective, which employs sets of positive and negative examples to align similar and repel dissimilar training data samples. However, similarity amongst training examples has a more continuous nature, thus calling for a more `non-binary' treatment. To address this, we propose a novel loss function called Continuously Weighted Contrastive Loss (CWCL) that employs a continuous measure of similarity. With CWCL, we seek to align the embedding space of one modality with another. Owing to the continuous nature of similarity in the proposed loss function, these models outperform existing methods for 0-shot transfer across multiple models, datasets and modalities. Particularly, we consider the modality pairs of image-text and speech-text and our models achieve 5-8% (absolute) improvement over previous state-of-the-art methods in 0-shot image classification and 20-30% (absolute) improvement in 0-shot speech-to-intent classification and keyword classification.
Improved Multi-Shot Diffusion-Weighted MRI with Zero-Shot Self-Supervised Learning Reconstruction
Aug 09, 2023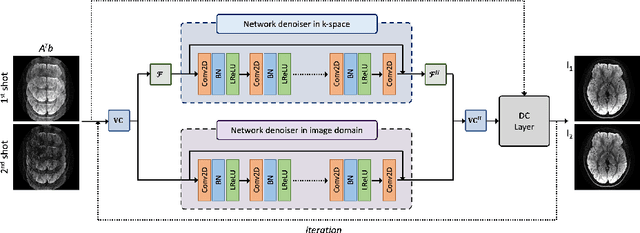
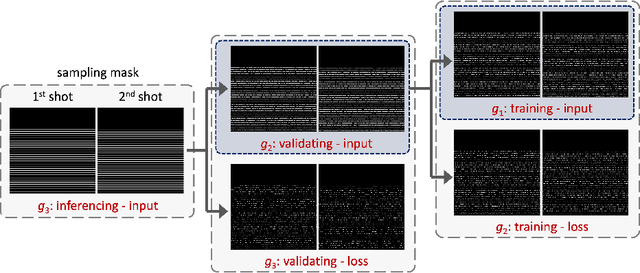
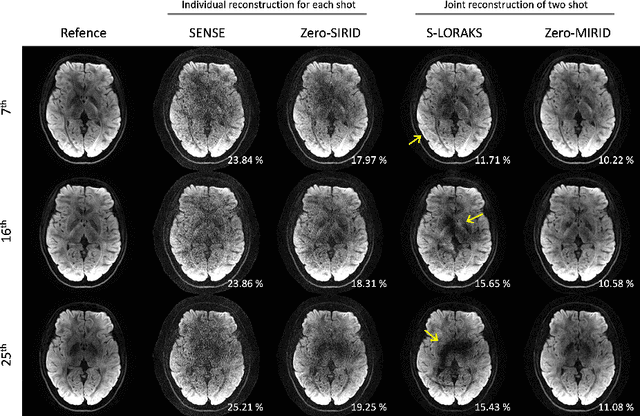

Abstract:Diffusion MRI is commonly performed using echo-planar imaging (EPI) due to its rapid acquisition time. However, the resolution of diffusion-weighted images is often limited by magnetic field inhomogeneity-related artifacts and blurring induced by T2- and T2*-relaxation effects. To address these limitations, multi-shot EPI (msEPI) combined with parallel imaging techniques is frequently employed. Nevertheless, reconstructing msEPI can be challenging due to phase variation between multiple shots. In this study, we introduce a novel msEPI reconstruction approach called zero-MIRID (zero-shot self-supervised learning of Multi-shot Image Reconstruction for Improved Diffusion MRI). This method jointly reconstructs msEPI data by incorporating deep learning-based image regularization techniques. The network incorporates CNN denoisers in both k- and image-spaces, while leveraging virtual coils to enhance image reconstruction conditioning. By employing a self-supervised learning technique and dividing sampled data into three groups, the proposed approach achieves superior results compared to the state-of-the-art parallel imaging method, as demonstrated in an in-vivo experiment.
Zero-DeepSub: Zero-Shot Deep Subspace Reconstruction for Rapid Multiparametric Quantitative MRI Using 3D-QALAS
Jul 04, 2023Abstract:Purpose: To develop and evaluate methods for 1) reconstructing 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) time-series images using a low-rank subspace method, which enables accurate and rapid T1 and T2 mapping, and 2) improving the fidelity of subspace QALAS by combining scan-specific deep-learning-based reconstruction and subspace modeling. Methods: A low-rank subspace method for 3D-QALAS (i.e., subspace QALAS) and zero-shot deep-learning subspace method (i.e., Zero-DeepSub) were proposed for rapid and high fidelity T1 and T2 mapping and time-resolved imaging using 3D-QALAS. Using an ISMRM/NIST system phantom, the accuracy of the T1 and T2 maps estimated using the proposed methods was evaluated by comparing them with reference techniques. The reconstruction performance of the proposed subspace QALAS using Zero-DeepSub was evaluated in vivo and compared with conventional QALAS at high reduction factors of up to 9-fold. Results: Phantom experiments showed that subspace QALAS had good linearity with respect to the reference methods while reducing biases compared to conventional QALAS, especially for T2 maps. Moreover, in vivo results demonstrated that subspace QALAS had better g-factor maps and could reduce voxel blurring, noise, and artifacts compared to conventional QALAS and showed robust performance at up to 9-fold acceleration with Zero-DeepSub, which enabled whole-brain T1, T2, and PD mapping at 1 mm isotropic resolution within 2 min of scan time. Conclusion: The proposed subspace QALAS along with Zero-DeepSub enabled high fidelity and rapid whole-brain multiparametric quantification and time-resolved imaging.
SSL-QALAS: Self-Supervised Learning for Rapid Multiparameter Estimation in Quantitative MRI Using 3D-QALAS
Feb 28, 2023Abstract:Purpose: To develop and evaluate a method for rapid estimation of multiparametric T1, T2, proton density (PD), and inversion efficiency (IE) maps from 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) measurements using self-supervised learning (SSL) without the need for an external dictionary. Methods: A SSL-based QALAS mapping method (SSL-QALAS) was developed for rapid and dictionary-free estimation of multiparametric maps from 3D-QALAS measurements. The accuracy of the reconstructed quantitative maps using dictionary matching and SSL-QALAS was evaluated by comparing the estimated T1 and T2 values with those obtained from the reference methods on an ISMRM/NIST phantom. The SSL-QALAS and the dictionary matching methods were also compared in vivo, and generalizability was evaluated by comparing the scan-specific, pre-trained, and transfer learning models. Results: Phantom experiments showed that both the dictionary matching and SSL-QALAS methods produced T1 and T2 estimates that had a strong linear agreement with the reference values in the ISMRM/NIST phantom. Further, SSL-QALAS showed similar performance with dictionary matching in reconstructing the T1, T2, PD, and IE maps on in vivo data. Rapid reconstruction of multiparametric maps was enabled by inferring the data using a pre-trained SSL-QALAS model within 10 s. Fast scan-specific tuning was also demonstrated by fine-tuning the pre-trained model with the target subject's data within 15 min. Conclusion: The proposed SSL-QALAS method enabled rapid reconstruction of multiparametric maps from 3D-QALAS measurements without an external dictionary or labeled ground-truth training data.
3D-EPI Blip-Up/Down Acquisition with CAIPI and Joint Hankel Structured Low-Rank Reconstruction for Rapid Distortion-Free High-Resolution T2* Mapping
Dec 01, 2022Abstract:Purpose: This work aims to develop a novel distortion-free 3D-EPI acquisition and image reconstruction technique for fast and robust, high-resolution, whole-brain imaging as well as quantitative T2* mapping. Methods: 3D-Blip-Up and -Down Acquisition (3D-BUDA) sequence is designed for both single- and multi-echo 3D GRE-EPI imaging using multiple shots with blip-up and -down readouts to encode B0 field map information. Complementary k-space coverage is achieved using controlled aliasing in parallel imaging (CAIPI) sampling across the shots. For image reconstruction, an iterative hard-thresholding algorithm is employed to minimize the cost function that combines field map information informed parallel imaging with the structured low-rank constraint for multi-shot 3D-BUDA data. Extending 3D-BUDA to multi-echo imaging permits T2* mapping. For this, we propose constructing a joint Hankel matrix along both echo and shot dimensions to improve the reconstruction. Results: Experimental results on in vivo multi-echo data demonstrate that, by performing joint reconstruction along with both echo and shot dimensions, reconstruction accuracy is improved compared to standard 3D-BUDA reconstruction. CAIPI sampling is further shown to enhance the image quality. For T2* mapping, T2* values from 3D-Joint-CAIPI-BUDA and reference multi-echo GRE are within limits of agreement as quantified by Bland-Altman analysis. Conclusions: The proposed technique enables rapid 3D distortion-free high-resolution imaging and T2* mapping. Specifically, 3D-BUDA enables 1-mm isotropic whole-brain imaging in 22 s at 3 T and 9 s on a 7 T scanner. The combination of multi-echo 3D-BUDA with CAIPI acquisition and joint reconstruction enables distortion-free whole-brain T2* mapping in 47 s at 1.1x1.1x1.0 mm3 resolution.
Time-efficient, High Resolution 3T Whole Brain Quantitative Relaxometry using 3D-QALAS with Wave-CAIPI Readouts
Nov 08, 2022



Abstract:Purpose: Volumetric, high resolution, quantitative mapping of brain tissues relaxation properties is hindered by long acquisition times and SNR challenges. This study, for the first time, combines the time efficient wave-CAIPI readouts into the 3D-QALAS acquisition scheme, enabling full brain quantitative T1, T2 and PD maps at 1.15 isotropic voxels in only 3 minutes. Methods: Wave-CAIPI readouts were embedded in the standard 3d-QALAS encoding scheme, enabling full brain quantitative parameter maps (T1, T2 and PD) at acceleration factors of R=3x2 with minimum SNR loss due to g-factor penalties. The quantitative maps using the accelerated protocol were quantitatively compared against those obtained from conventional 3D-QALAS sequence using GRAPPA acceleration of R=2 in the ISMRM NIST phantom, and ten healthy volunteers. To show the feasibility of the proposed methods in clinical settings, the accelerated wave-CAIPI 3D-QALAS sequence was also employed in pediatric patients undergoing clinical MRI examinations. Results: When tested in both the ISMRM/NIST phantom and 7 healthy volunteers, the quantitative maps using the accelerated protocol showed excellent agreement against those obtained from conventional 3D-QALAS at R=2. Conclusion: 3D-QALAS enhanced with wave-CAIPI readouts enables time-efficient, full brain quantitative T1, T2 and PD mapping at 1.15 in 3 minutes at R=3x2 acceleration. When tested on the NIST phantom and 7 healthy volunteers, the quantitative maps obtained from the accelerated wave-CAIPI 3D-QALAS protocol showed very similar values to those obtained from the standard 3D-QALAS (R=2) protocol, alluding to the robustness and reliability of the proposed methods. This study also shows that the accelerated protocol can be effectively employed in pediatric patient populations, making high-quality high-resolution full brain quantitative imaging feasible in clinical settings.
Non-Contrastive Self-supervised Learning for Utterance-Level Information Extraction from Speech
Aug 10, 2022



Abstract:In recent studies, self-supervised pre-trained models tend to outperform supervised pre-trained models in transfer learning. In particular, self-supervised learning (SSL) of utterance-level speech representation can be used in speech applications that require discriminative representation of consistent attributes within an utterance: speaker, language, emotion, and age. Existing frame-level self-supervised speech representation, e.g., wav2vec, can be used as utterance-level representation with pooling, but the models are usually large. There are also SSL techniques to learn utterance-level representation. One of the most successful is a contrastive method, which requires negative sampling: selecting alternative samples to contrast with the current sample (anchor). However, this does not ensure that all the negative samples belong to classes different from the anchor class without labels. This paper applies a non-contrastive self-supervised method to learn utterance-level embeddings. We adapted DIstillation with NO labels (DINO) from computer vision to speech. Unlike contrastive methods, DINO does not require negative sampling. We compared DINO to x-vector trained in a supervised manner. When transferred to down-stream tasks (speaker verification, speech emotion recognition (SER), and Alzheimer's disease detection), DINO outperformed x-vector. We studied the influence of several aspects during transfer learning such as dividing the fine-tuning process into steps, chunk lengths, or augmentation. During fine-tuning, tuning the last affine layers first and then the whole network surpassed fine-tuning all at once. Using shorter chunk lengths, although they generate more diverse inputs, did not necessarily improve performance, implying speech segments at least with a specific length are required for better performance per application. Augmentation was helpful in SER.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge