Anastasia Yendiki
PRIME: Phase Reversed Interleaved Multi-Echo acquisition enables highly accelerated distortion-free diffusion MRI
Sep 11, 2024


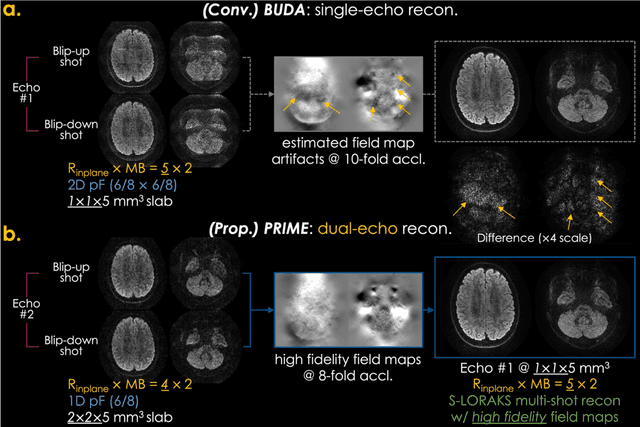
Abstract:Purpose: To develop and evaluate a new pulse sequence for highly accelerated distortion-free diffusion MRI (dMRI) by inserting an additional echo without prolonging TR, when generalized slice dithered enhanced resolution (gSlider) radiofrequency encoding is used for volumetric acquisition. Methods: A phase-reversed interleaved multi-echo acquisition (PRIME) was developed for rapid, high-resolution, and distortion-free dMRI, which includes two echoes where the first echo is for target diffusion-weighted imaging (DWI) acquisition with high-resolution and the second echo is acquired with either 1) lower-resolution for high-fidelity field map estimation, or 2) matching resolution to enable efficient diffusion relaxometry acquisitions. The sequence was evaluated on in vivo data acquired from healthy volunteers on clinical and Connectome 2.0 scanners. Results: In vivo experiments demonstrated that 1) high in-plane acceleration (Rin-plane of 5-fold with 2D partial Fourier) was achieved using the high-fidelity field maps estimated from the second echo, which was made at a lower resolution/acceleration to increase its SNR while matching the effective echo spacing of the first readout, 2) high-resolution diffusion relaxometry parameters were estimated from dual-echo PRIME data using a white matter model of multi-TE spherical mean technique (MTE-SMT), and 3) high-fidelity mesoscale DWI at 550 um isotropic resolution could be obtained in vivo by capitalizing on the high-performance gradients of the Connectome 2.0 scanner. Conclusion: The proposed PRIME sequence enabled highly accelerated, high-resolution, and distortion-free dMRI using an additional echo without prolonging scan time when gSlider encoding is utilized.
Domain-agnostic segmentation of thalamic nuclei from joint structural and diffusion MRI
May 05, 2023Abstract:The human thalamus is a highly connected subcortical grey-matter structure within the brain. It comprises dozens of nuclei with different function and connectivity, which are affected differently by disease. For this reason, there is growing interest in studying the thalamic nuclei in vivo with MRI. Tools are available to segment the thalamus from 1 mm T1 scans, but the contrast of the lateral and internal boundaries is too faint to produce reliable segmentations. Some tools have attempted to incorporate information from diffusion MRI in the segmentation to refine these boundaries, but do not generalise well across diffusion MRI acquisitions. Here we present the first CNN that can segment thalamic nuclei from T1 and diffusion data of any resolution without retraining or fine tuning. Our method builds on a public histological atlas of the thalamic nuclei and silver standard segmentations on high-quality diffusion data obtained with a recent Bayesian adaptive segmentation tool. We combine these with an approximate degradation model for fast domain randomisation during training. Our CNN produces a segmentation at 0.7 mm isotropic resolution, irrespective of the resolution of the input. Moreover, it uses a parsimonious model of the diffusion signal at each voxel (fractional anisotropy and principal eigenvector) that is compatible with virtually any set of directions and b-values, including huge amounts of legacy data. We show results of our proposed method on three heterogeneous datasets acquired on dozens of different scanners. An implementation of the method is publicly available at https://freesurfer.net/fswiki/ThalamicNucleiDTI.
Constrained self-supervised method with temporal ensembling for fiber bundle detection on anatomic tracing data
Aug 06, 2022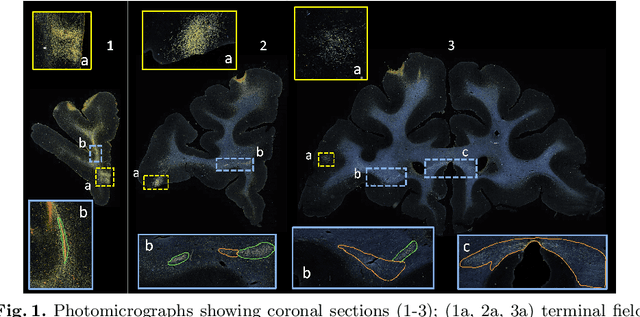
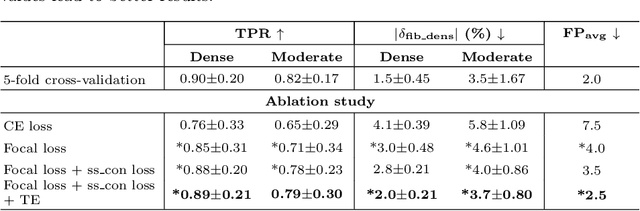
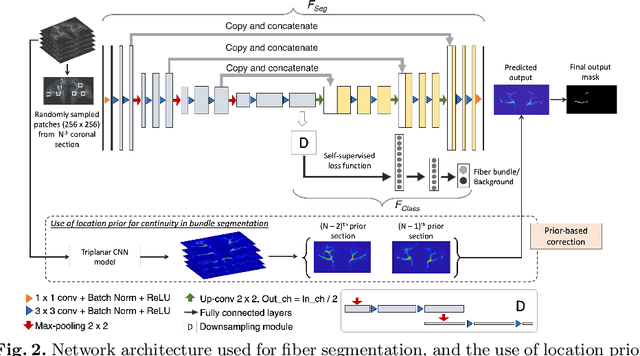
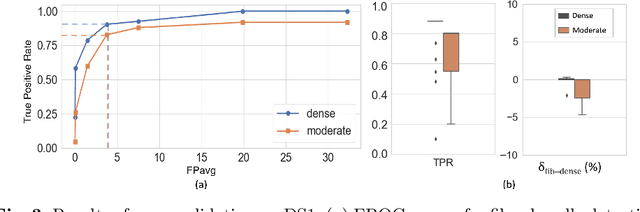
Abstract:Anatomic tracing data provides detailed information on brain circuitry essential for addressing some of the common errors in diffusion MRI tractography. However, automated detection of fiber bundles on tracing data is challenging due to sectioning distortions, presence of noise and artifacts and intensity/contrast variations. In this work, we propose a deep learning method with a self-supervised loss function that takes anatomy-based constraints into account for accurate segmentation of fiber bundles on the tracer sections from macaque brains. Also, given the limited availability of manual labels, we use a semi-supervised training technique for efficiently using unlabeled data to improve the performance, and location constraints for further reduction of false positives. Evaluation of our method on unseen sections from a different macaque yields promising results with a true positive rate of ~0.90. The code for our method is available at https://github.com/v-sundaresan/fiberbundle_seg_tracing.
Learning Anatomical Segmentations for Tractography from Diffusion MRI
Sep 09, 2020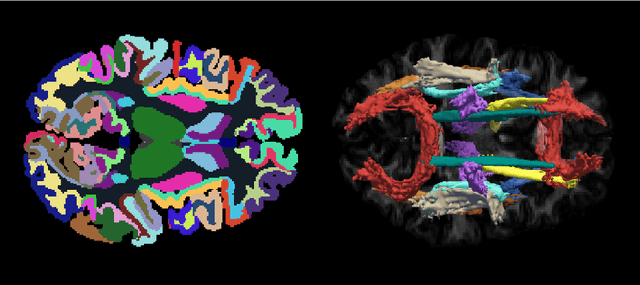
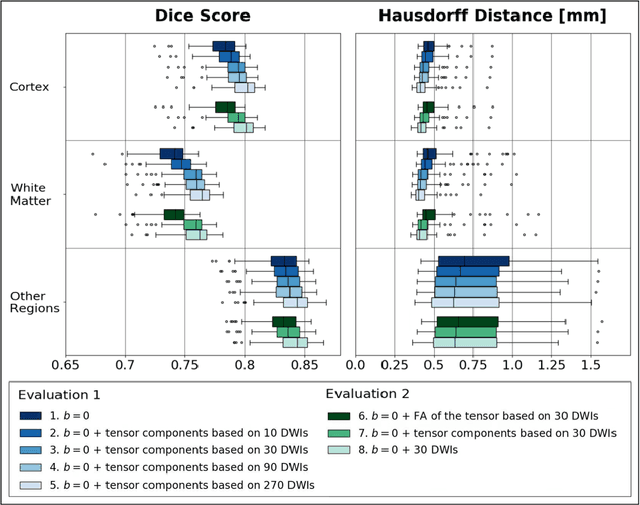
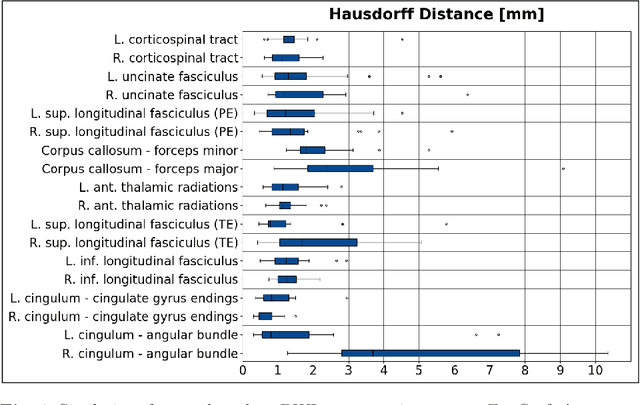
Abstract:Deep learning approaches for diffusion MRI have so far focused primarily on voxel-based segmentation of lesions or white-matter fiber tracts. A drawback of representing tracts as volumetric labels, rather than sets of streamlines, is that it precludes point-wise analyses of microstructural or geometric features along a tract. Traditional tractography pipelines, which do allow such analyses, can benefit from detailed whole-brain segmentations to guide tract reconstruction. Here, we introduce fast, deep learning-based segmentation of 170 anatomical regions directly on diffusion-weighted MR images, removing the dependency of conventional segmentation methods on T 1-weighted images and slow pre-processing pipelines. Working natively in diffusion space avoids non-linear distortions and registration errors across modalities, as well as interpolation artifacts. We demonstrate consistent segmentation results between 0 .70 and 0 .87 Dice depending on the tissue type. We investigate various combinations of diffusion-derived inputs and show generalization across different numbers of gradient directions. Finally, integrating our approach to provide anatomical priors for tractography pipelines, such as TRACULA, removes hours of pre-processing time and permits processing even in the absence of high-quality T 1-weighted scans, without degrading the quality of the resulting tract estimates.
Joint inference on structural and diffusion MRI for sequence-adaptive Bayesian segmentation of thalamic nuclei with probabilistic atlases
Mar 11, 2019
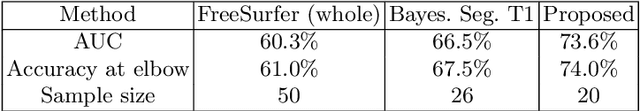
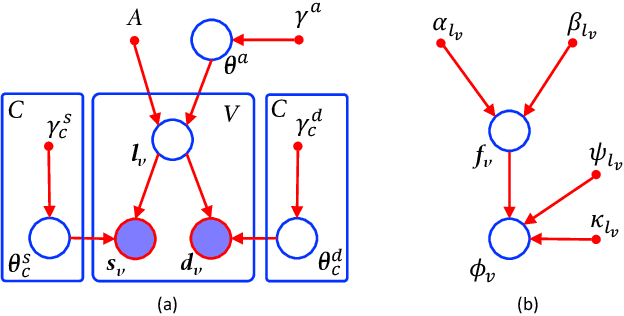
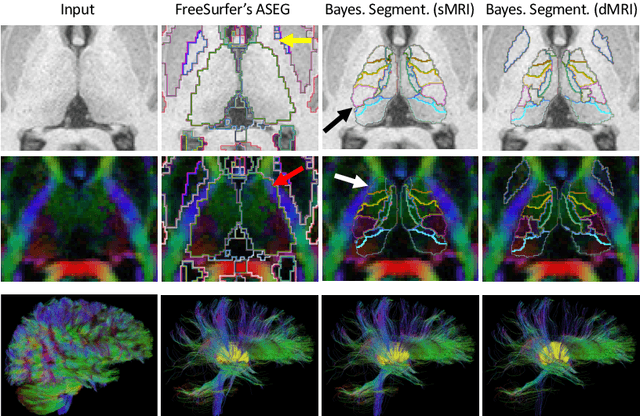
Abstract:Segmentation of structural and diffusion MRI (sMRI/dMRI) is usually performed independently in neuroimaging pipelines. However, some brain structures (e.g., globus pallidus, thalamus and its nuclei) can be extracted more accurately by fusing the two modalities. Following the framework of Bayesian segmentation with probabilistic atlases and unsupervised appearance modeling, we present here a novel algorithm to jointly segment multi-modal sMRI/dMRI data. We propose a hierarchical likelihood term for the dMRI defined on the unit ball, which combines the Beta and Dimroth-Scheidegger-Watson distributions to model the data at each voxel. This term is integrated with a mixture of Gaussians for the sMRI data, such that the resulting joint unsupervised likelihood enables the analysis of multi-modal scans acquired with any type of MRI contrast, b-values, or number of directions, which enables wide applicability. We also propose an inference algorithm to estimate the maximum-a-posteriori model parameters from input images, and to compute the most likely segmentation. Using a recently published atlas derived from histology, we apply our method to thalamic nuclei segmentation on two datasets: HCP (state of the art) and ADNI (legacy) - producing lower sample sizes than Bayesian segmentation with sMRI alone.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge