David Kügler
Regression is all you need for medical image translation
May 06, 2025Abstract:The acquisition of information-rich images within a limited time budget is crucial in medical imaging. Medical image translation (MIT) can help enhance and supplement existing datasets by generating synthetic images from acquired data. While Generative Adversarial Nets (GANs) and Diffusion Models (DMs) have achieved remarkable success in natural image generation, their benefits - creativity and image realism - do not necessarily transfer to medical applications where highly accurate anatomical information is required. In fact, the imitation of acquisition noise or content hallucination hinder clinical utility. Here, we introduce YODA (You Only Denoise once - or Average), a novel 2.5D diffusion-based framework for volumetric MIT. YODA unites diffusion and regression paradigms to produce realistic or noise-free outputs. Furthermore, we propose Expectation-Approximation (ExpA) DM sampling, which draws inspiration from MRI signal averaging. ExpA-sampling suppresses generated noise and, thus, eliminates noise from biasing the evaluation of image quality. Through extensive experiments on four diverse multi-modal datasets - comprising multi-contrast brain MRI and pelvic MRI-CT - we show that diffusion and regression sampling yield similar results in practice. As such, the computational overhead of diffusion sampling does not provide systematic benefits in medical information translation. Building on these insights, we demonstrate that YODA outperforms several state-of-the-art GAN and DM methods. Notably, YODA-generated images are shown to be interchangeable with, or even superior to, physical acquisitions for several downstream tasks. Our findings challenge the presumed advantages of DMs in MIT and pave the way for the practical application of MIT in medical imaging.
Regression s all you need for medical image translation
May 04, 2025Abstract:The acquisition of information-rich images within a limited time budget is crucial in medical imaging. Medical image translation (MIT) can help enhance and supplement existing datasets by generating synthetic images from acquired data. While Generative Adversarial Nets (GANs) and Diffusion Models (DMs) have achieved remarkable success in natural image generation, their benefits - creativity and image realism - do not necessarily transfer to medical applications where highly accurate anatomical information is required. In fact, the imitation of acquisition noise or content hallucination hinder clinical utility. Here, we introduce YODA (You Only Denoise once - or Average), a novel 2.5D diffusion-based framework for volumetric MIT. YODA unites diffusion and regression paradigms to produce realistic or noise-free outputs. Furthermore, we propose Expectation-Approximation (ExpA) DM sampling, which draws inspiration from MRI signal averaging. ExpA-sampling suppresses generated noise and, thus, eliminates noise from biasing the evaluation of image quality. Through extensive experiments on four diverse multi-modal datasets - comprising multi-contrast brain MRI and pelvic MRI-CT - we show that diffusion and regression sampling yield similar results in practice. As such, the computational overhead of diffusion sampling does not provide systematic benefits in medical information translation. Building on these insights, we demonstrate that YODA outperforms several state-of-the-art GAN and DM methods. Notably, YODA-generated images are shown to be interchangeable with, or even superior to, physical acquisitions for several downstream tasks. Our findings challenge the presumed advantages of DMs in MIT and pave the way for the practical application of MIT in medical imaging.
VINNA for Neonates -- Orientation Independence through Latent Augmentations
Nov 29, 2023Abstract:Fast and accurate segmentation of neonatal brain images is highly desired to better understand and detect changes during development and disease. Yet, the limited availability of ground truth datasets, lack of standardized acquisition protocols, and wide variations of head positioning pose challenges for method development. A few automated image analysis pipelines exist for newborn brain MRI segmentation, but they often rely on time-consuming procedures and require resampling to a common resolution, subject to loss of information due to interpolation and down-sampling. Without registration and image resampling, variations with respect to head positions and voxel resolutions have to be addressed differently. In deep-learning, external augmentations are traditionally used to artificially expand the representation of spatial variability, increasing the training dataset size and robustness. However, these transformations in the image space still require resampling, reducing accuracy specifically in the context of label interpolation. We recently introduced the concept of resolution-independence with the Voxel-size Independent Neural Network framework, VINN. Here, we extend this concept by additionally shifting all rigid-transforms into the network architecture with a four degree of freedom (4-DOF) transform module, enabling resolution-aware internal augmentations (VINNA). In this work we show that VINNA (i) significantly outperforms state-of-the-art external augmentation approaches, (ii) effectively addresses the head variations present specifically in newborn datasets, and (iii) retains high segmentation accuracy across a range of resolutions (0.5-1.0 mm). The 4-DOF transform module is a powerful, general approach to implement spatial augmentation without requiring image or label interpolation. The specific network application to newborns will be made publicly available as VINNA4neonates.
FastSurfer-HypVINN: Automated sub-segmentation of the hypothalamus and adjacent structures on high-resolutional brain MRI
Aug 24, 2023Abstract:The hypothalamus plays a crucial role in the regulation of a broad range of physiological, behavioural, and cognitive functions. However, despite its importance, only a few small-scale neuroimaging studies have investigated its substructures, likely due to the lack of fully automated segmentation tools to address scalability and reproducibility issues of manual segmentation. While the only previous attempt to automatically sub-segment the hypothalamus with a neural network showed promise for 1.0 mm isotropic T1-weighted (T1w) MRI, there is a need for an automated tool to sub-segment also high-resolutional (HiRes) MR scans, as they are becoming widely available, and include structural detail also from multi-modal MRI. We, therefore, introduce a novel, fast, and fully automated deep learning method named HypVINN for sub-segmentation of the hypothalamus and adjacent structures on 0.8 mm isotropic T1w and T2w brain MR images that is robust to missing modalities. We extensively validate our model with respect to segmentation accuracy, generalizability, in-session test-retest reliability, and sensitivity to replicate hypothalamic volume effects (e.g. sex-differences). The proposed method exhibits high segmentation performance both for standalone T1w images as well as for T1w/T2w image pairs. Even with the additional capability to accept flexible inputs, our model matches or exceeds the performance of state-of-the-art methods with fixed inputs. We, further, demonstrate the generalizability of our method in experiments with 1.0 mm MR scans from both the Rhineland Study and the UK Biobank. Finally, HypVINN can perform the segmentation in less than a minute (GPU) and will be available in the open source FastSurfer neuroimaging software suite, offering a validated, efficient, and scalable solution for evaluating imaging-derived phenotypes of the hypothalamus.
Estimating Head Motion from MR-Images
Feb 28, 2023Abstract:Head motion is an omnipresent confounder of magnetic resonance image (MRI) analyses as it systematically affects morphometric measurements, even when visual quality control is performed. In order to estimate subtle head motion, that remains undetected by experts, we introduce a deep learning method to predict in-scanner head motion directly from T1-weighted (T1w), T2-weighted (T2w) and fluid-attenuated inversion recovery (FLAIR) images using motion estimates from an in-scanner depth camera as ground truth. Since we work with data from compliant healthy participants of the Rhineland Study, head motion and resulting imaging artifacts are less prevalent than in most clinical cohorts and more difficult to detect. Our method demonstrates improved performance compared to state-of-the-art motion estimation methods and can quantify drift and respiration movement independently. Finally, on unseen data, our predictions preserve the known, significant correlation with age.
Identifying and Combating Bias in Segmentation Networks by leveraging multiple resolutions
Jun 29, 2022



Abstract:Exploration of bias has significant impact on the transparency and applicability of deep learning pipelines in medical settings, yet is so far woefully understudied. In this paper, we consider two separate groups for which training data is only available at differing image resolutions. For group H, available images and labels are at the preferred high resolution while for group L only deprecated lower resolution data exist. We analyse how this resolution-bias in the data distribution propagates to systematically biased predictions for group L at higher resolutions. Our results demonstrate that single-resolution training settings result in significant loss of volumetric group differences that translate to erroneous segmentations as measured by DSC and subsequent classification failures on the low resolution group. We further explore how training data across resolutions can be used to combat this systematic bias. Specifically, we investigate the effect of image resampling, scale augmentation and resolution independence and demonstrate that biases can effectively be reduced with multi-resolution approaches.
FastSurferVINN: Building Resolution-Independence into Deep Learning Segmentation Methods -- A Solution for HighRes Brain MRI
Dec 17, 2021

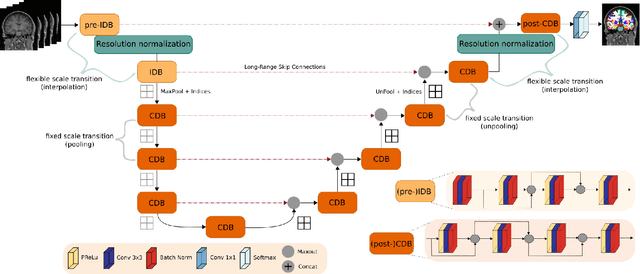

Abstract:Leading neuroimaging studies have pushed 3T MRI acquisition resolutions below 1.0 mm for improved structure definition and morphometry. Yet, only few, time-intensive automated image analysis pipelines have been validated for high-resolution (HiRes) settings. Efficient deep learning approaches, on the other hand, rarely support more than one fixed resolution (usually 1.0 mm). Furthermore, the lack of a standard submillimeter resolution as well as limited availability of diverse HiRes data with sufficient coverage of scanner, age, diseases, or genetic variance poses additional, unsolved challenges for training HiRes networks. Incorporating resolution-independence into deep learning-based segmentation, i.e., the ability to segment images at their native resolution across a range of different voxel sizes, promises to overcome these challenges, yet no such approach currently exists. We now fill this gap by introducing a Voxelsize Independent Neural Network (VINN) for resolution-independent segmentation tasks and present FastSurferVINN, which (i) establishes and implements resolution-independence for deep learning as the first method simultaneously supporting 0.7-1.0 mm whole brain segmentation, (ii) significantly outperforms state-of-the-art methods across resolutions, and (iii) mitigates the data imbalance problem present in HiRes datasets. Overall, internal resolution-independence mutually benefits both HiRes and 1.0 mm MRI segmentation. With our rigorously validated FastSurferVINN we distribute a rapid tool for morphometric neuroimage analysis. The VINN architecture, furthermore, represents an efficient resolution-independent segmentation method for wider application
Learning Anatomical Segmentations for Tractography from Diffusion MRI
Sep 09, 2020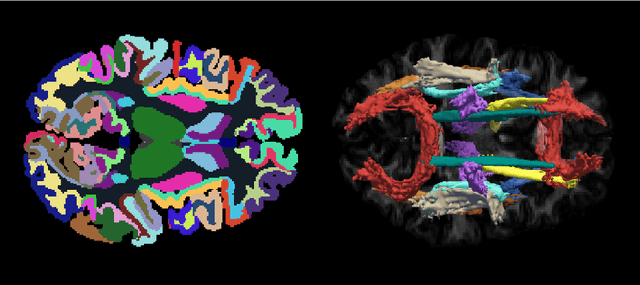
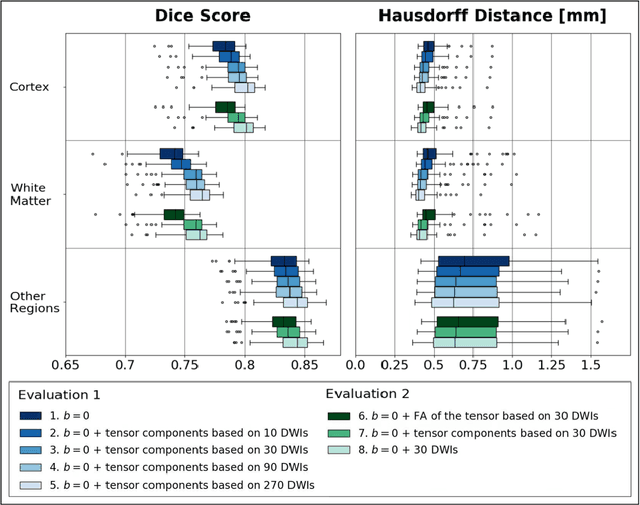
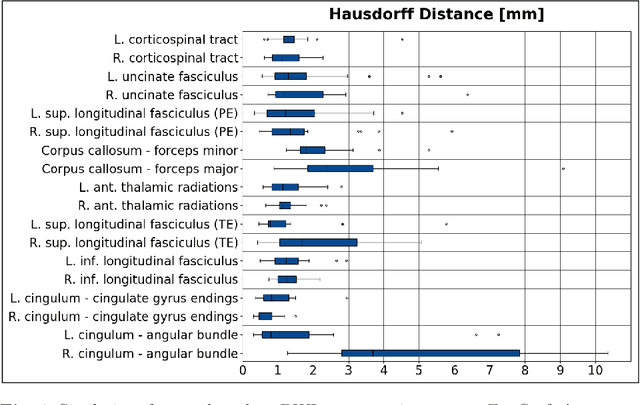
Abstract:Deep learning approaches for diffusion MRI have so far focused primarily on voxel-based segmentation of lesions or white-matter fiber tracts. A drawback of representing tracts as volumetric labels, rather than sets of streamlines, is that it precludes point-wise analyses of microstructural or geometric features along a tract. Traditional tractography pipelines, which do allow such analyses, can benefit from detailed whole-brain segmentations to guide tract reconstruction. Here, we introduce fast, deep learning-based segmentation of 170 anatomical regions directly on diffusion-weighted MR images, removing the dependency of conventional segmentation methods on T 1-weighted images and slow pre-processing pipelines. Working natively in diffusion space avoids non-linear distortions and registration errors across modalities, as well as interpolation artifacts. We demonstrate consistent segmentation results between 0 .70 and 0 .87 Dice depending on the tissue type. We investigate various combinations of diffusion-derived inputs and show generalization across different numbers of gradient directions. Finally, integrating our approach to provide anatomical priors for tractography pipelines, such as TRACULA, removes hours of pre-processing time and permits processing even in the absence of high-quality T 1-weighted scans, without degrading the quality of the resulting tract estimates.
AutoSNAP: Automatically Learning Neural Architectures for Instrument Pose Estimation
Jun 26, 2020



Abstract:Despite recent successes, the advances in Deep Learning have not yet been fully translated to Computer Assisted Intervention (CAI) problems such as pose estimation of surgical instruments. Currently, neural architectures for classification and segmentation tasks are adopted ignoring significant discrepancies between CAI and these tasks. We propose an automatic framework (AutoSNAP) for instrument pose estimation problems, which discovers and learns the architectures for neural networks. We introduce 1)~an efficient testing environment for pose estimation, 2)~a powerful architecture representation based on novel Symbolic Neural Architecture Patterns (SNAPs), and 3)~an optimization of the architecture using an efficient search scheme. Using AutoSNAP, we discover an improved architecture (SNAPNet) which outperforms both the hand-engineered i3PosNet and the state-of-the-art architecture search method DARTS.
Exploring Adversarial Examples: Patterns of One-Pixel Attacks
Jun 25, 2018



Abstract:Failure cases of black-box deep learning, e.g. adversarial examples, might have severe consequences in healthcare. Yet such failures are mostly studied in the context of real-world images with calibrated attacks. To demystify the adversarial examples, rigorous studies need to be designed. Unfortunately, complexity of the medical images hinders such study design directly from the medical images. We hypothesize that adversarial examples might result from the incorrect mapping of image space to the low dimensional generation manifold by deep networks. To test the hypothesis, we simplify a complex medical problem namely pose estimation of surgical tools into its barest form. An analytical decision boundary and exhaustive search of the one-pixel attack across multiple image dimensions let us localize the regions of frequent successful one-pixel attacks at the image space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge