Lilla Zöllei
Unified Brain Surface and Volume Registration
Dec 22, 2025Abstract:Accurate registration of brain MRI scans is fundamental for cross-subject analysis in neuroscientific studies. This involves aligning both the cortical surface of the brain and the interior volume. Traditional methods treat volumetric and surface-based registration separately, which often leads to inconsistencies that limit downstream analyses. We propose a deep learning framework, NeurAlign, that registers $3$D brain MRI images by jointly aligning both cortical and subcortical regions through a unified volume-and-surface-based representation. Our approach leverages an intermediate spherical coordinate space to bridge anatomical surface topology with volumetric anatomy, enabling consistent and anatomically accurate alignment. By integrating spherical registration into the learning, our method ensures geometric coherence between volume and surface domains. In a series of experiments on both in-domain and out-of-domain datasets, our method consistently outperforms both classical and machine learning-based registration methods -- improving the Dice score by up to 7 points while maintaining regular deformation fields. Additionally, it is orders of magnitude faster than the standard method for this task, and is simpler to use because it requires no additional inputs beyond an MRI scan. With its superior accuracy, fast inference, and ease of use, NeurAlign sets a new standard for joint cortical and subcortical registration.
Search Wide, Focus Deep: Automated Fetal Brain Extraction with Sparse Training Data
Oct 29, 2024



Abstract:Automated fetal brain extraction from full-uterus MRI is a challenging task due to variable head sizes, orientations, complex anatomy, and prevalent artifacts. While deep-learning (DL) models trained on synthetic images have been successful in adult brain extraction, adapting these networks for fetal MRI is difficult due to the sparsity of labeled data, leading to increased false-positive predictions. To address this challenge, we propose a test-time strategy that reduces false positives in networks trained on sparse, synthetic labels. The approach uses a breadth-fine search (BFS) to identify a subvolume likely to contain the fetal brain, followed by a deep-focused sliding window (DFS) search to refine the extraction, pooling predictions to minimize false positives. We train models at different window sizes using synthetic images derived from a small number of fetal brain label maps, augmented with random geometric shapes. Each model is trained on diverse head positions and scales, including cases with partial or no brain tissue. Our framework matches state-of-the-art brain extraction methods on clinical HASTE scans of third-trimester fetuses and exceeds them by up to 5\% in terms of Dice in the second trimester as well as EPI scans across both trimesters. Our results demonstrate the utility of a sliding-window approach and combining predictions from several models trained on synthetic images, for improving brain-extraction accuracy by progressively refining regions of interest and minimizing the risk of missing brain mask slices or misidentifying other tissues as brain.
Boosting Skull-Stripping Performance for Pediatric Brain Images
Feb 26, 2024



Abstract:Skull-stripping is the removal of background and non-brain anatomical features from brain images. While many skull-stripping tools exist, few target pediatric populations. With the emergence of multi-institutional pediatric data acquisition efforts to broaden the understanding of perinatal brain development, it is essential to develop robust and well-tested tools ready for the relevant data processing. However, the broad range of neuroanatomical variation in the developing brain, combined with additional challenges such as high motion levels, as well as shoulder and chest signal in the images, leaves many adult-specific tools ill-suited for pediatric skull-stripping. Building on an existing framework for robust and accurate skull-stripping, we propose developmental SynthStrip (d-SynthStrip), a skull-stripping model tailored to pediatric images. This framework exposes networks to highly variable images synthesized from label maps. Our model substantially outperforms pediatric baselines across scan types and age cohorts. In addition, the <1-minute runtime of our tool compares favorably to the fastest baselines. We distribute our model at https://w3id.org/synthstrip.
VINNA for Neonates -- Orientation Independence through Latent Augmentations
Nov 29, 2023Abstract:Fast and accurate segmentation of neonatal brain images is highly desired to better understand and detect changes during development and disease. Yet, the limited availability of ground truth datasets, lack of standardized acquisition protocols, and wide variations of head positioning pose challenges for method development. A few automated image analysis pipelines exist for newborn brain MRI segmentation, but they often rely on time-consuming procedures and require resampling to a common resolution, subject to loss of information due to interpolation and down-sampling. Without registration and image resampling, variations with respect to head positions and voxel resolutions have to be addressed differently. In deep-learning, external augmentations are traditionally used to artificially expand the representation of spatial variability, increasing the training dataset size and robustness. However, these transformations in the image space still require resampling, reducing accuracy specifically in the context of label interpolation. We recently introduced the concept of resolution-independence with the Voxel-size Independent Neural Network framework, VINN. Here, we extend this concept by additionally shifting all rigid-transforms into the network architecture with a four degree of freedom (4-DOF) transform module, enabling resolution-aware internal augmentations (VINNA). In this work we show that VINNA (i) significantly outperforms state-of-the-art external augmentation approaches, (ii) effectively addresses the head variations present specifically in newborn datasets, and (iii) retains high segmentation accuracy across a range of resolutions (0.5-1.0 mm). The 4-DOF transform module is a powerful, general approach to implement spatial augmentation without requiring image or label interpolation. The specific network application to newborns will be made publicly available as VINNA4neonates.
Automating Whole Brain Histology to MRI Registration: Implementation of a Computational Pipeline
May 22, 2019



Abstract:Although the latest advances in MRI technology have allowed the acquisition of higher resolution images, reliable delineation of cytoarchitectural or subcortical nuclei boundaries is not possible. As a result, histological images are still required to identify the exact limits of neuroanatomical structures. However, histological processing is associated with tissue distortion and fixation artifacts, which prevent a direct comparison between the two modalities. Our group has previously proposed a histological procedure based on celloidin embedding that reduces the amount of artifacts and yields high quality whole brain histological slices. Celloidin embedded tissue, nevertheless, still bears distortions that must be corrected. We propose a computational pipeline designed to semi-automatically process the celloidin embedded histology and register them to their MRI counterparts. In this paper we report the accuracy of our pipeline in two whole brain volumes from the Brain Bank of the Brazilian Aging Brain Study Group (BBBABSG). Results were assessed by comparison of manual segmentations from two experts in both MRIs and the registered histological volumes. The two whole brain histology/MRI datasets were successfully registered using minimal user interaction. We also point to possible improvements based on recent implementations that could be added to this pipeline, potentially allowing for higher precision and further performance gains.
Fast Infant MRI Skullstripping with Multiview 2D Convolutional Neural Networks
Apr 27, 2019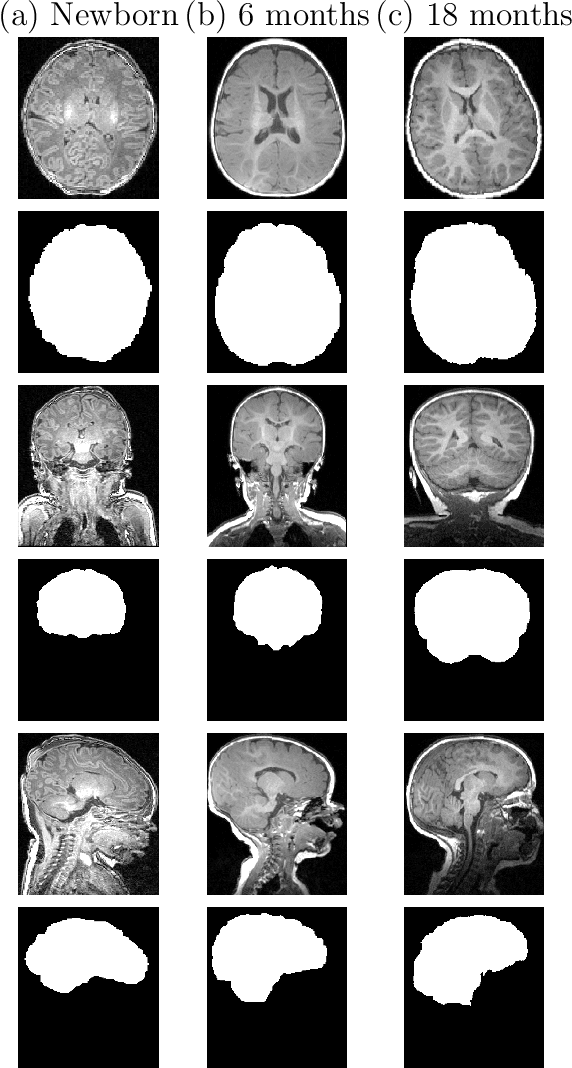

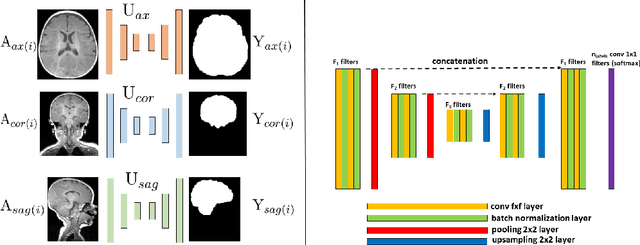
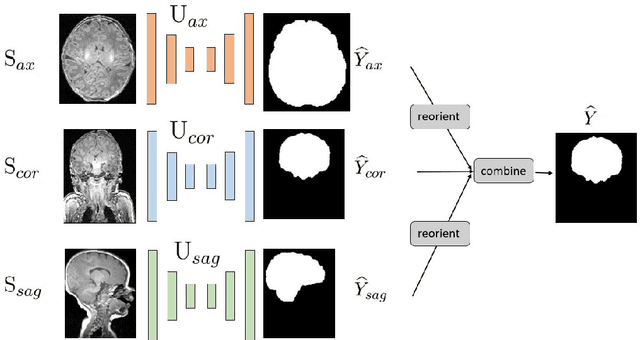
Abstract:Skullstripping is defined as the task of segmenting brain tissue from a full head magnetic resonance image~(MRI). It is a critical component in neuroimage processing pipelines. Downstream deformable registration and whole brain segmentation performance is highly dependent on accurate skullstripping. Skullstripping is an especially challenging task for infant~(age range 0--18 months) head MRI images due to the significant size and shape variability of the head and the brain in that age range. Infant brain tissue development also changes the $T_1$-weighted image contrast over time, making consistent skullstripping a difficult task. Existing tools for adult brain MRI skullstripping are ill equipped to handle these variations and a specialized infant MRI skullstripping algorithm is necessary. In this paper, we describe a supervised skullstripping algorithm that utilizes three trained fully convolutional neural networks~(CNN), each of which segments 2D $T_1$-weighted slices in axial, coronal, and sagittal views respectively. The three probabilistic segmentations in the three views are linearly fused and thresholded to produce a final brain mask. We compared our method to existing adult and infant skullstripping algorithms and showed significant improvement based on Dice overlap metric~(average Dice of 0.97) with a manually labeled ground truth data set. Label fusion experiments on multiple, unlabeled data sets show that our method is consistent and has fewer failure modes. In addition, our method is computationally very fast with a run time of 30 seconds per image on NVidia P40/P100/Quadro 4000 GPUs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge