Amod Jog
Fast Infant MRI Skullstripping with Multiview 2D Convolutional Neural Networks
Apr 27, 2019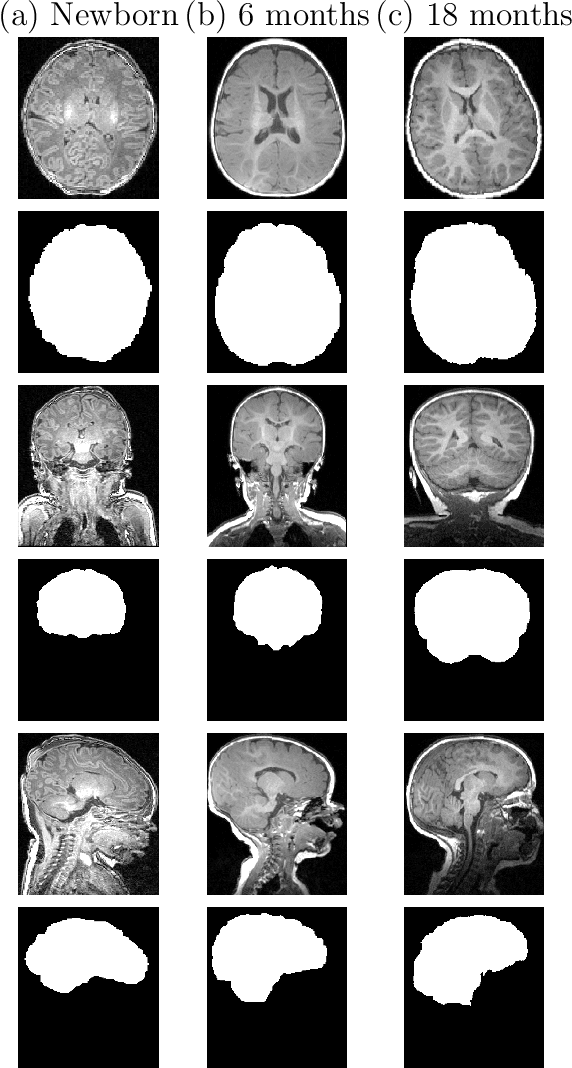

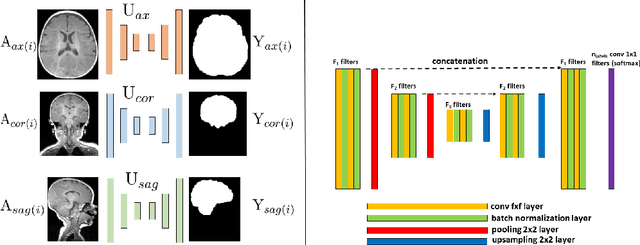
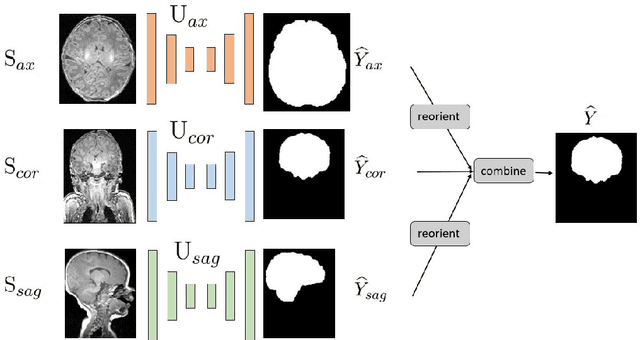
Abstract:Skullstripping is defined as the task of segmenting brain tissue from a full head magnetic resonance image~(MRI). It is a critical component in neuroimage processing pipelines. Downstream deformable registration and whole brain segmentation performance is highly dependent on accurate skullstripping. Skullstripping is an especially challenging task for infant~(age range 0--18 months) head MRI images due to the significant size and shape variability of the head and the brain in that age range. Infant brain tissue development also changes the $T_1$-weighted image contrast over time, making consistent skullstripping a difficult task. Existing tools for adult brain MRI skullstripping are ill equipped to handle these variations and a specialized infant MRI skullstripping algorithm is necessary. In this paper, we describe a supervised skullstripping algorithm that utilizes three trained fully convolutional neural networks~(CNN), each of which segments 2D $T_1$-weighted slices in axial, coronal, and sagittal views respectively. The three probabilistic segmentations in the three views are linearly fused and thresholded to produce a final brain mask. We compared our method to existing adult and infant skullstripping algorithms and showed significant improvement based on Dice overlap metric~(average Dice of 0.97) with a manually labeled ground truth data set. Label fusion experiments on multiple, unlabeled data sets show that our method is consistent and has fewer failure modes. In addition, our method is computationally very fast with a run time of 30 seconds per image on NVidia P40/P100/Quadro 4000 GPUs.
PSACNN: Pulse Sequence Adaptive Fast Whole Brain Segmentation
Jan 21, 2019



Abstract:With the advent of convolutional neural networks~(CNN), supervised learning methods are increasingly being used for whole brain segmentation. However, a large, manually annotated training dataset of labeled brain images required to train such supervised methods is frequently difficult to obtain or create. In addition, existing training datasets are generally acquired with a homogeneous magnetic resonance imaging~(MRI) acquisition protocol. CNNs trained on such datasets are unable to generalize on test data with different acquisition protocols. Modern neuroimaging studies and clinical trials are necessarily multi-center initiatives with a wide variety of acquisition protocols. Despite stringent protocol harmonization practices, it is very difficult to standardize the gamut of MRI imaging parameters across scanners, field strengths, receive coils etc., that affect image contrast. In this paper we propose a CNN-based segmentation algorithm that, in addition to being highly accurate and fast, is also resilient to variation in the input acquisition. Our approach relies on building approximate forward models of pulse sequences that produce a typical test image. For a given pulse sequence, we use its forward model to generate plausible, synthetic training examples that appear as if they were acquired in a scanner with that pulse sequence. Sampling over a wide variety of pulse sequences results in a wide variety of augmented training examples that help build an image contrast invariant model. Our method trains a single CNN that can segment input MRI images with acquisition parameters as disparate as $T_1$-weighted and $T_2$-weighted contrasts with only $T_1$-weighted training data. The segmentations generated are highly accurate with state-of-the-art results~(overall Dice overlap$=0.94$), with a fast run time~($\approx$ 45 seconds), and consistent across a wide range of acquisition protocols.
Pulse Sequence Resilient Fast Brain Segmentation
Jul 30, 2018



Abstract:Accurate automatic segmentation of brain anatomy from $T_1$-weighted~($T_1$-w) magnetic resonance images~(MRI) has been a computationally intensive bottleneck in neuroimaging pipelines, with state-of-the-art results obtained by unsupervised intensity modeling-based methods and multi-atlas registration and label fusion. With the advent of powerful supervised convolutional neural networks~(CNN)-based learning algorithms, it is now possible to produce a high quality brain segmentation within seconds. However, the very supervised nature of these methods makes it difficult to generalize them on data different from what they have been trained on. Modern neuroimaging studies are necessarily multi-center initiatives with a wide variety of acquisition protocols. Despite stringent protocol harmonization practices, it is not possible to standardize the whole gamut of MRI imaging parameters across scanners, field strengths, receive coils etc., that affect image contrast. In this paper we propose a CNN-based segmentation algorithm that, in addition to being highly accurate and fast, is also resilient to variation in the input $T_1$-w acquisition. Our approach relies on building approximate forward models of $T_1$-w pulse sequences that produce a typical test image. We use the forward models to augment the training data with test data specific training examples. These augmented data can be used to update and/or build a more robust segmentation model that is more attuned to the test data imaging properties. Our method generates highly accurate, state-of-the-art segmentation results~(overall Dice overlap=0.94), within seconds and is consistent across a wide-range of protocols.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge