Yohan Jun
Explainable AI for Accelerated Microstructure Imaging: A SHAP-Guided Protocol on the Connectome 2.0 scanner
Sep 11, 2025Abstract:The diffusion MRI Neurite Exchange Imaging model offers a promising framework for probing gray matter microstructure by estimating parameters such as compartment sizes, diffusivities, and inter-compartmental water exchange time. However, existing protocols require long scan times. This study proposes a reduced acquisition scheme for the Connectome 2.0 scanner that preserves model accuracy while substantially shortening scan duration. We developed a data-driven framework using explainable artificial intelligence with a guided recursive feature elimination strategy to identify an optimal 8-feature subset from a 15-feature protocol. The performance of this optimized protocol was validated in vivo and benchmarked against the full acquisition and alternative reduction strategies. Parameter accuracy, preservation of anatomical contrast, and test-retest reproducibility were assessed. The reduced protocol yielded parameter estimates and cortical maps comparable to the full protocol, with low estimation errors in synthetic data and minimal impact on test-retest variability. Compared to theory-driven and heuristic reduction schemes, the optimized protocol demonstrated superior robustness, reducing the deviation in water exchange time estimates by over two-fold. In conclusion, this hybrid optimization framework enables viable imaging of neurite exchange in 14 minutes without loss of parameter fidelity. This approach supports the broader application of exchange-sensitive diffusion magnetic resonance imaging in neuroscience and clinical research, and offers a generalizable method for designing efficient acquisition protocols in biophysical parameter mapping.
MIMOSA: Multi-parametric Imaging using Multiple-echoes with Optimized Simultaneous Acquisition for highly-efficient quantitative MRI
Aug 13, 2025



Abstract:Purpose: To develop a new sequence, MIMOSA, for highly-efficient T1, T2, T2*, proton density (PD), and source separation quantitative susceptibility mapping (QSM). Methods: MIMOSA was developed based on 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) by combining 3D turbo Fast Low Angle Shot (FLASH) and multi-echo gradient echo acquisition modules with a spiral-like Cartesian trajectory to facilitate highly-efficient acquisition. Simulations were performed to optimize the sequence. Multi-contrast/-slice zero-shot self-supervised learning algorithm was employed for reconstruction. The accuracy of quantitative mapping was assessed by comparing MIMOSA with 3D-QALAS and reference techniques in both ISMRM/NIST phantom and in-vivo experiments. MIMOSA's acceleration capability was assessed at R = 3.3, 6.5, and 11.8 in in-vivo experiments, with repeatability assessed through scan-rescan studies. Beyond the 3T experiments, mesoscale quantitative mapping was performed at 750 um isotropic resolution at 7T. Results: Simulations demonstrated that MIMOSA achieved improved parameter estimation accuracy compared to 3D-QALAS. Phantom experiments indicated that MIMOSA exhibited better agreement with the reference techniques than 3D-QALAS. In-vivo experiments demonstrated that an acceleration factor of up to R = 11.8-fold can be achieved while preserving parameter estimation accuracy, with intra-class correlation coefficients of 0.998 (T1), 0.973 (T2), 0.947 (T2*), 0.992 (QSM), 0.987 (paramagnetic susceptibility), and 0.977 (diamagnetic susceptibility) in scan-rescan studies. Whole-brain T1, T2, T2*, PD, source separation QSM were obtained with 1 mm isotropic resolution in 3 min at 3T and 750 um isotropic resolution in 13 min at 7T. Conclusion: MIMOSA demonstrated potential for highly-efficient multi-parametric mapping.
A Tutorial on MRI Reconstruction: From Modern Methods to Clinical Implications
Jul 22, 2025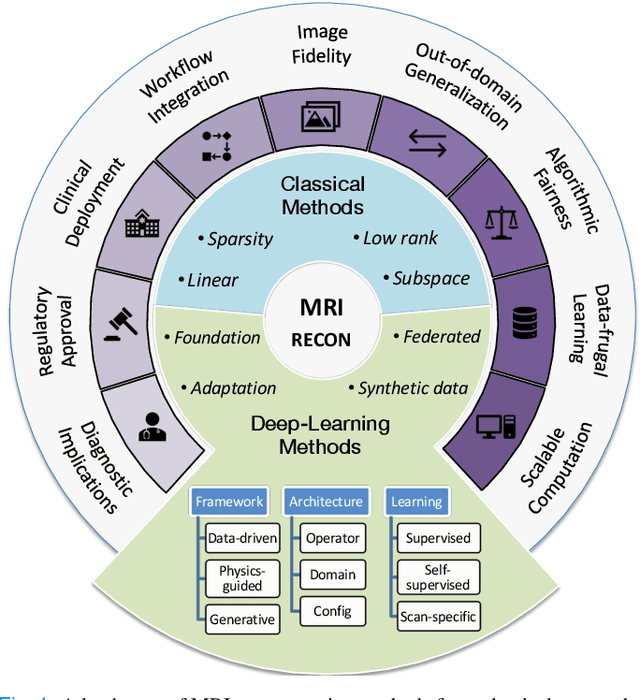
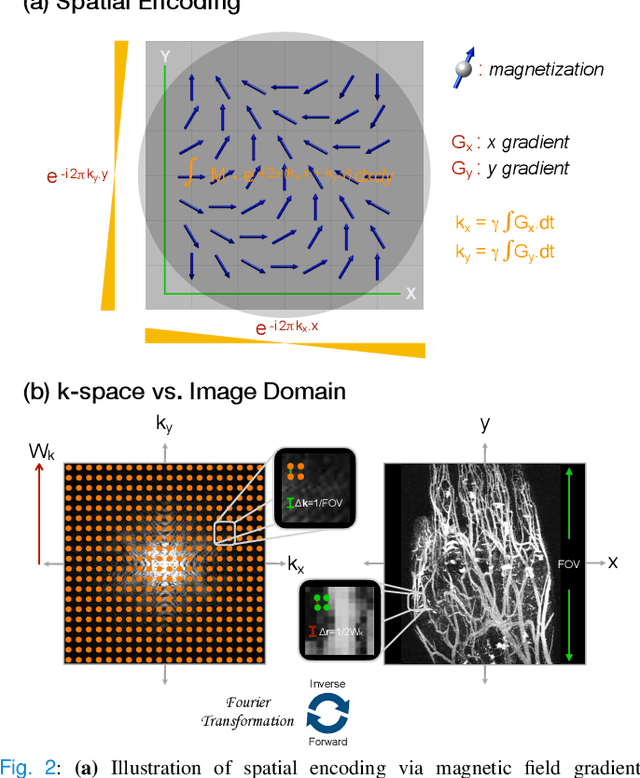
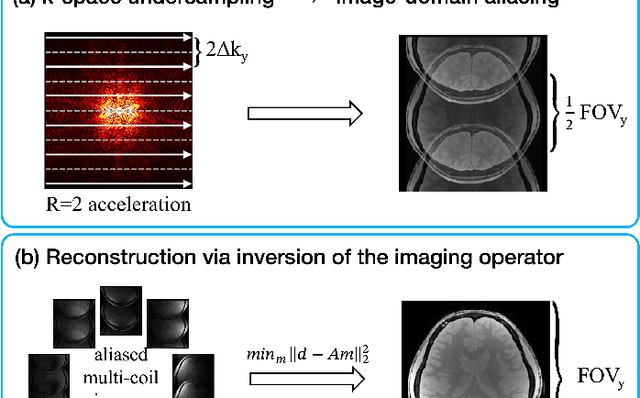
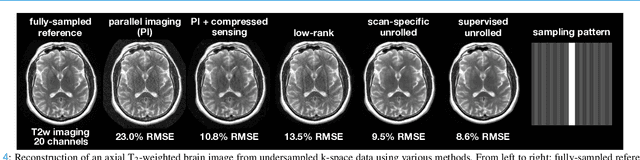
Abstract:MRI is an indispensable clinical tool, offering a rich variety of tissue contrasts to support broad diagnostic and research applications. Clinical exams routinely acquire multiple structural sequences that provide complementary information for differential diagnosis, while research protocols often incorporate advanced functional, diffusion, spectroscopic, and relaxometry sequences to capture multidimensional insights into tissue structure and composition. However, these capabilities come at the cost of prolonged scan times, which reduce patient throughput, increase susceptibility to motion artifacts, and may require trade-offs in image quality or diagnostic scope. Over the last two decades, advances in image reconstruction algorithms--alongside improvements in hardware and pulse sequence design--have made it possible to accelerate acquisitions while preserving diagnostic quality. Central to this progress is the ability to incorporate prior information to regularize the solutions to the reconstruction problem. In this tutorial, we overview the basics of MRI reconstruction and highlight state-of-the-art approaches, beginning with classical methods that rely on explicit hand-crafted priors, and then turning to deep learning methods that leverage a combination of learned and crafted priors to further push the performance envelope. We also explore the translational aspects and eventual clinical implications of these methods. We conclude by discussing future directions to address remaining challenges in MRI reconstruction. The tutorial is accompanied by a Python toolbox (https://github.com/tutorial-MRI-recon/tutorial) to demonstrate select methods discussed in the article.
PRIME: Phase Reversed Interleaved Multi-Echo acquisition enables highly accelerated distortion-free diffusion MRI
Sep 11, 2024


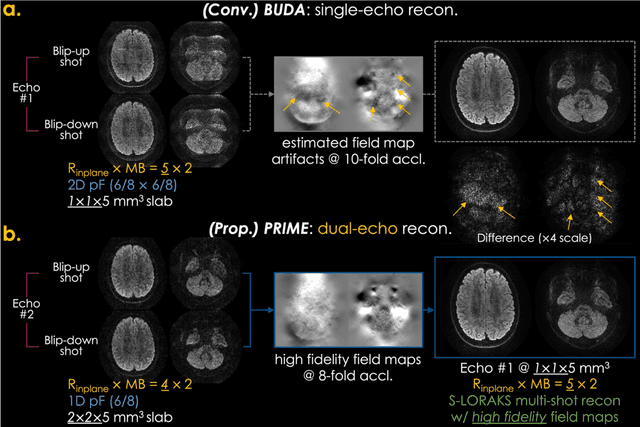
Abstract:Purpose: To develop and evaluate a new pulse sequence for highly accelerated distortion-free diffusion MRI (dMRI) by inserting an additional echo without prolonging TR, when generalized slice dithered enhanced resolution (gSlider) radiofrequency encoding is used for volumetric acquisition. Methods: A phase-reversed interleaved multi-echo acquisition (PRIME) was developed for rapid, high-resolution, and distortion-free dMRI, which includes two echoes where the first echo is for target diffusion-weighted imaging (DWI) acquisition with high-resolution and the second echo is acquired with either 1) lower-resolution for high-fidelity field map estimation, or 2) matching resolution to enable efficient diffusion relaxometry acquisitions. The sequence was evaluated on in vivo data acquired from healthy volunteers on clinical and Connectome 2.0 scanners. Results: In vivo experiments demonstrated that 1) high in-plane acceleration (Rin-plane of 5-fold with 2D partial Fourier) was achieved using the high-fidelity field maps estimated from the second echo, which was made at a lower resolution/acceleration to increase its SNR while matching the effective echo spacing of the first readout, 2) high-resolution diffusion relaxometry parameters were estimated from dual-echo PRIME data using a white matter model of multi-TE spherical mean technique (MTE-SMT), and 3) high-fidelity mesoscale DWI at 550 um isotropic resolution could be obtained in vivo by capitalizing on the high-performance gradients of the Connectome 2.0 scanner. Conclusion: The proposed PRIME sequence enabled highly accelerated, high-resolution, and distortion-free dMRI using an additional echo without prolonging scan time when gSlider encoding is utilized.
NLCG-Net: A Model-Based Zero-Shot Learning Framework for Undersampled Quantitative MRI Reconstruction
Jan 22, 2024Abstract:Typical quantitative MRI (qMRI) methods estimate parameter maps after image reconstructing, which is prone to biases and error propagation. We propose a Nonlinear Conjugate Gradient (NLCG) optimizer for model-based T2/T1 estimation, which incorporates U-Net regularization trained in a scan-specific manner. This end-to-end method directly estimates qMRI maps from undersampled k-space data using mono-exponential signal modeling with zero-shot scan-specific neural network regularization to enable high fidelity T1 and T2 mapping. T2 and T1 mapping results demonstrate the ability of the proposed NLCG-Net to improve estimation quality compared to subspace reconstruction at high accelerations.
Improved Multi-Shot Diffusion-Weighted MRI with Zero-Shot Self-Supervised Learning Reconstruction
Aug 09, 2023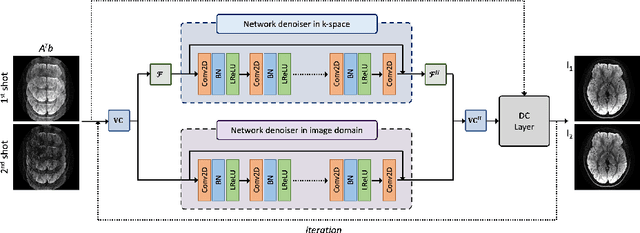
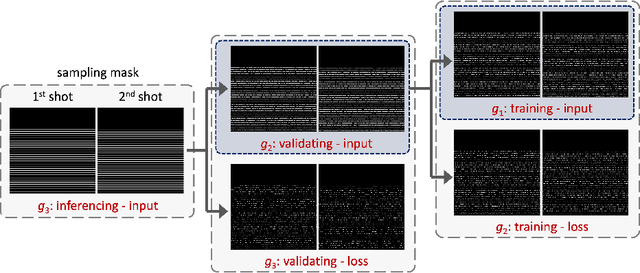
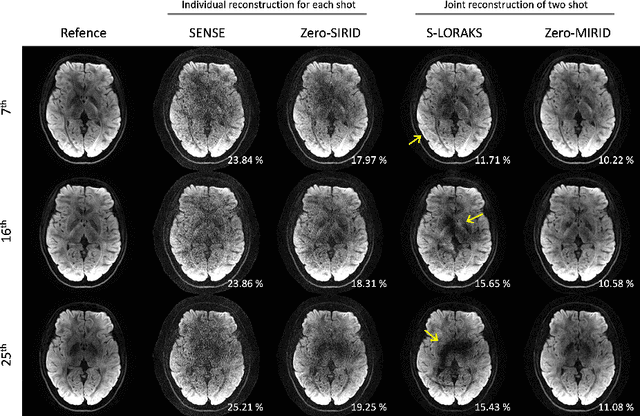

Abstract:Diffusion MRI is commonly performed using echo-planar imaging (EPI) due to its rapid acquisition time. However, the resolution of diffusion-weighted images is often limited by magnetic field inhomogeneity-related artifacts and blurring induced by T2- and T2*-relaxation effects. To address these limitations, multi-shot EPI (msEPI) combined with parallel imaging techniques is frequently employed. Nevertheless, reconstructing msEPI can be challenging due to phase variation between multiple shots. In this study, we introduce a novel msEPI reconstruction approach called zero-MIRID (zero-shot self-supervised learning of Multi-shot Image Reconstruction for Improved Diffusion MRI). This method jointly reconstructs msEPI data by incorporating deep learning-based image regularization techniques. The network incorporates CNN denoisers in both k- and image-spaces, while leveraging virtual coils to enhance image reconstruction conditioning. By employing a self-supervised learning technique and dividing sampled data into three groups, the proposed approach achieves superior results compared to the state-of-the-art parallel imaging method, as demonstrated in an in-vivo experiment.
Zero-DeepSub: Zero-Shot Deep Subspace Reconstruction for Rapid Multiparametric Quantitative MRI Using 3D-QALAS
Jul 04, 2023Abstract:Purpose: To develop and evaluate methods for 1) reconstructing 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) time-series images using a low-rank subspace method, which enables accurate and rapid T1 and T2 mapping, and 2) improving the fidelity of subspace QALAS by combining scan-specific deep-learning-based reconstruction and subspace modeling. Methods: A low-rank subspace method for 3D-QALAS (i.e., subspace QALAS) and zero-shot deep-learning subspace method (i.e., Zero-DeepSub) were proposed for rapid and high fidelity T1 and T2 mapping and time-resolved imaging using 3D-QALAS. Using an ISMRM/NIST system phantom, the accuracy of the T1 and T2 maps estimated using the proposed methods was evaluated by comparing them with reference techniques. The reconstruction performance of the proposed subspace QALAS using Zero-DeepSub was evaluated in vivo and compared with conventional QALAS at high reduction factors of up to 9-fold. Results: Phantom experiments showed that subspace QALAS had good linearity with respect to the reference methods while reducing biases compared to conventional QALAS, especially for T2 maps. Moreover, in vivo results demonstrated that subspace QALAS had better g-factor maps and could reduce voxel blurring, noise, and artifacts compared to conventional QALAS and showed robust performance at up to 9-fold acceleration with Zero-DeepSub, which enabled whole-brain T1, T2, and PD mapping at 1 mm isotropic resolution within 2 min of scan time. Conclusion: The proposed subspace QALAS along with Zero-DeepSub enabled high fidelity and rapid whole-brain multiparametric quantification and time-resolved imaging.
SDC-UDA: Volumetric Unsupervised Domain Adaptation Framework for Slice-Direction Continuous Cross-Modality Medical Image Segmentation
May 18, 2023Abstract:Recent advances in deep learning-based medical image segmentation studies achieve nearly human-level performance in fully supervised manner. However, acquiring pixel-level expert annotations is extremely expensive and laborious in medical imaging fields. Unsupervised domain adaptation (UDA) can alleviate this problem, which makes it possible to use annotated data in one imaging modality to train a network that can successfully perform segmentation on target imaging modality with no labels. In this work, we propose SDC-UDA, a simple yet effective volumetric UDA framework for slice-direction continuous cross-modality medical image segmentation which combines intra- and inter-slice self-attentive image translation, uncertainty-constrained pseudo-label refinement, and volumetric self-training. Our method is distinguished from previous methods on UDA for medical image segmentation in that it can obtain continuous segmentation in the slice direction, thereby ensuring higher accuracy and potential in clinical practice. We validate SDC-UDA with multiple publicly available cross-modality medical image segmentation datasets and achieve state-of-the-art segmentation performance, not to mention the superior slice-direction continuity of prediction compared to previous studies.
SSL-QALAS: Self-Supervised Learning for Rapid Multiparameter Estimation in Quantitative MRI Using 3D-QALAS
Feb 28, 2023Abstract:Purpose: To develop and evaluate a method for rapid estimation of multiparametric T1, T2, proton density (PD), and inversion efficiency (IE) maps from 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) measurements using self-supervised learning (SSL) without the need for an external dictionary. Methods: A SSL-based QALAS mapping method (SSL-QALAS) was developed for rapid and dictionary-free estimation of multiparametric maps from 3D-QALAS measurements. The accuracy of the reconstructed quantitative maps using dictionary matching and SSL-QALAS was evaluated by comparing the estimated T1 and T2 values with those obtained from the reference methods on an ISMRM/NIST phantom. The SSL-QALAS and the dictionary matching methods were also compared in vivo, and generalizability was evaluated by comparing the scan-specific, pre-trained, and transfer learning models. Results: Phantom experiments showed that both the dictionary matching and SSL-QALAS methods produced T1 and T2 estimates that had a strong linear agreement with the reference values in the ISMRM/NIST phantom. Further, SSL-QALAS showed similar performance with dictionary matching in reconstructing the T1, T2, PD, and IE maps on in vivo data. Rapid reconstruction of multiparametric maps was enabled by inferring the data using a pre-trained SSL-QALAS model within 10 s. Fast scan-specific tuning was also demonstrated by fine-tuning the pre-trained model with the target subject's data within 15 min. Conclusion: The proposed SSL-QALAS method enabled rapid reconstruction of multiparametric maps from 3D-QALAS measurements without an external dictionary or labeled ground-truth training data.
COSMOS: Cross-Modality Unsupervised Domain Adaptation for 3D Medical Image Segmentation based on Target-aware Domain Translation and Iterative Self-Training
Mar 30, 2022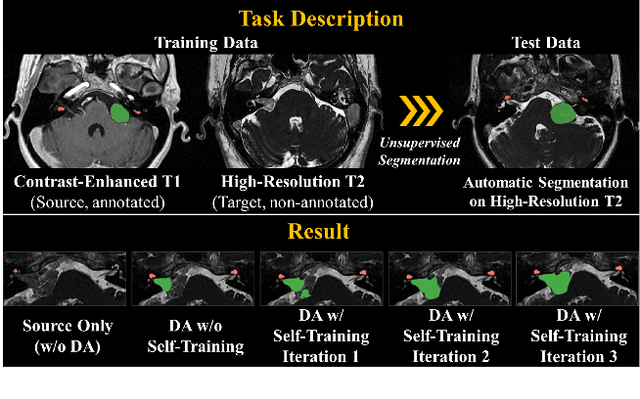
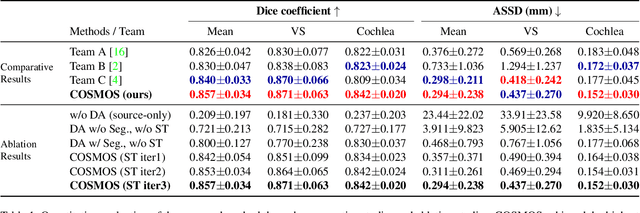
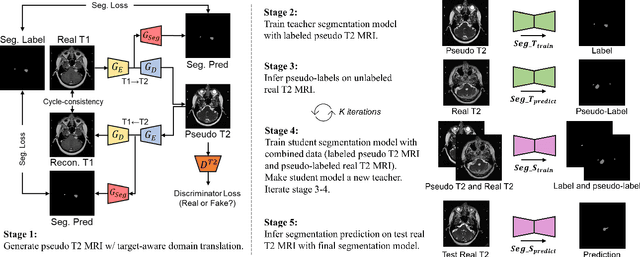
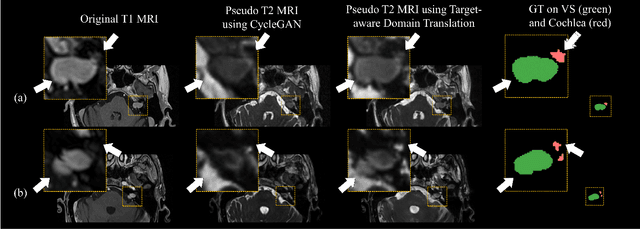
Abstract:Recent advances in deep learning-based medical image segmentation studies achieve nearly human-level performance when in fully supervised condition. However, acquiring pixel-level expert annotations is extremely expensive and laborious in medical imaging fields. Unsupervised domain adaptation can alleviate this problem, which makes it possible to use annotated data in one imaging modality to train a network that can successfully perform segmentation on target imaging modality with no labels. In this work, we propose a self-training based unsupervised domain adaptation framework for 3D medical image segmentation named COSMOS and validate it with automatic segmentation of Vestibular Schwannoma (VS) and cochlea on high-resolution T2 Magnetic Resonance Images (MRI). Our target-aware contrast conversion network translates source domain annotated T1 MRI to pseudo T2 MRI to enable segmentation training on target domain, while preserving important anatomical features of interest in the converted images. Iterative self-training is followed to incorporate unlabeled data to training and incrementally improve the quality of pseudo-labels, thereby leading to improved performance of segmentation. COSMOS won the 1\textsuperscript{st} place in the Cross-Modality Domain Adaptation (crossMoDA) challenge held in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). It achieves mean Dice score and Average Symmetric Surface Distance of 0.871(0.063) and 0.437(0.270) for VS, and 0.842(0.020) and 0.152(0.030) for cochlea.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge