Mahmoud Mostapha
Scale-Cascaded Diffusion Models for Super-Resolution in Medical Imaging
Jan 30, 2026Abstract:Diffusion models have been increasingly used as strong generative priors for solving inverse problems such as super-resolution in medical imaging. However, these approaches typically utilize a diffusion prior trained at a single scale, ignoring the hierarchical scale structure of image data. In this work, we propose to decompose images into Laplacian pyramid scales and train separate diffusion priors for each frequency band. We then develop an algorithm to perform super-resolution that utilizes these priors to progressively refine reconstructions across different scales. Evaluated on brain, knee, and prostate MRI data, our approach both improves perceptual quality over baselines and reduces inference time through smaller coarse-scale networks. Our framework unifies multiscale reconstruction and diffusion priors for medical image super-resolution.
Investigating the Feasibility of Patch-based Inference for Generalized Diffusion Priors in Inverse Problems for Medical Images
Jan 25, 2025Abstract:Plug-and-play approaches to solving inverse problems such as restoration and super-resolution have recently benefited from Diffusion-based generative priors for natural as well as medical images. However, solutions often use the standard albeit computationally intensive route of training and inferring with the whole image on the diffusion prior. While patch-based approaches to evaluating diffusion priors in plug-and-play methods have received some interest, they remain an open area of study. In this work, we explore the feasibility of the usage of patches for training and inference of a diffusion prior on MRI images. We explore the minor adaptation necessary for artifact avoidance, the performance and the efficiency of memory usage of patch-based methods as well as the adaptability of whole image training to patch-based evaluation - evaluating across multiple plug-and-play methods, tasks and datasets.
State-of-the-Art Machine Learning MRI Reconstruction in 2020: Results of the Second fastMRI Challenge
Dec 28, 2020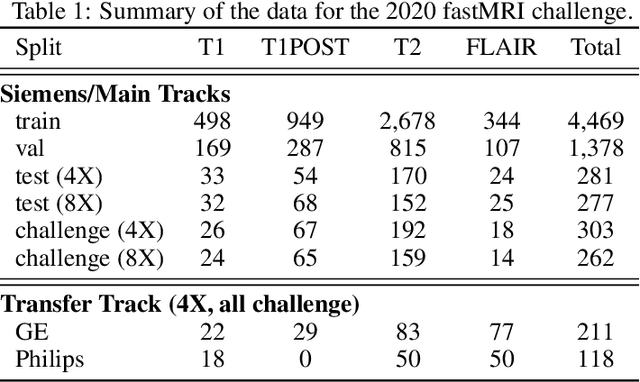
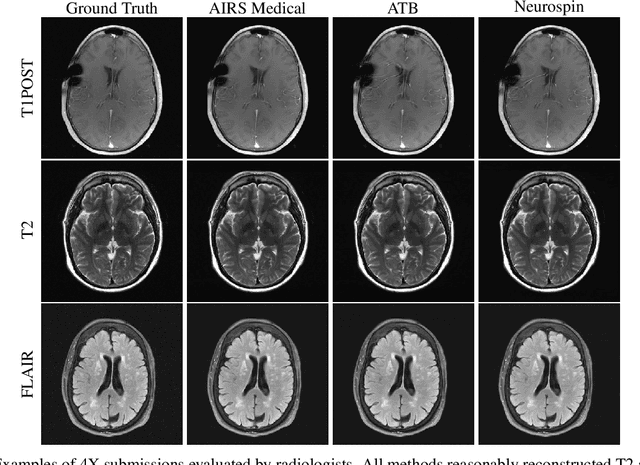
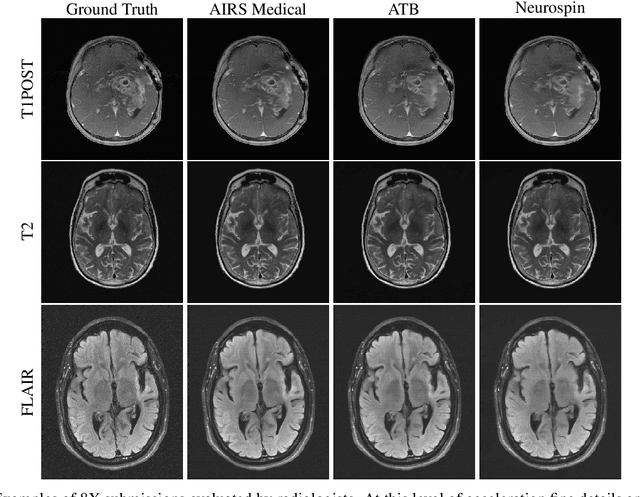
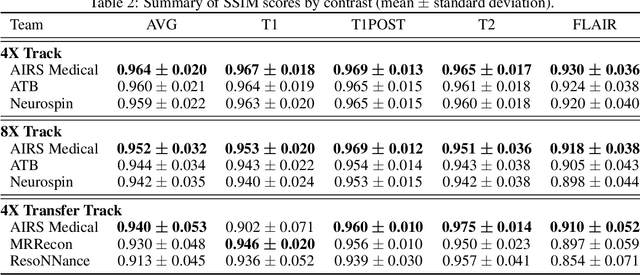
Abstract:Accelerating MRI scans is one of the principal outstanding problems in the MRI research community. Towards this goal, we hosted the second fastMRI competition targeted towards reconstructing MR images with subsampled k-space data. We provided participants with data from 7,299 clinical brain scans (de-identified via a HIPAA-compliant procedure by NYU Langone Health), holding back the fully-sampled data from 894 of these scans for challenge evaluation purposes. In contrast to the 2019 challenge, we focused our radiologist evaluations on pathological assessment in brain images. We also debuted a new Transfer track that required participants to submit models evaluated on MRI scanners from outside the training set. We received 19 submissions from eight different groups. Results showed one team scoring best in both SSIM scores and qualitative radiologist evaluations. We also performed analysis on alternative metrics to mitigate the effects of background noise and collected feedback from the participants to inform future challenges. Lastly, we identify common failure modes across the submissions, highlighting areas of need for future research in the MRI reconstruction community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge