Martina Bocchetta
High-resolution segmentations of the hypothalamus and its subregions for training of segmentation models
Jun 27, 2024Abstract:Segmentation of brain structures on magnetic resonance imaging (MRI) is a highly relevant neuroimaging topic, as it is a prerequisite for different analyses such as volumetry or shape analysis. Automated segmentation facilitates the study of brain structures in larger cohorts when compared with manual segmentation, which is time-consuming. However, the development of most automated methods relies on large and manually annotated datasets, which limits the generalizability of these methods. Recently, new techniques using synthetic images have emerged, reducing the need for manual annotation. Here we provide HELM, Hypothalamic ex vivo Label Maps, a dataset composed of label maps built from publicly available ultra-high resolution ex vivo MRI from 10 whole hemispheres, which can be used to develop segmentation methods using synthetic data. The label maps are obtained with a combination of manual labels for the hypothalamic regions and automated segmentations for the rest of the brain, and mirrored to simulate entire brains. We also provide the pre-processed ex vivo scans, as this dataset can support future projects to include other structures after these are manually segmented.
H-SynEx: Using synthetic images and ultra-high resolution ex vivo MRI for hypothalamus subregion segmentation
Jan 30, 2024



Abstract:Purpose: To develop a method for automated segmentation of hypothalamus subregions informed by ultra-high resolution ex vivo magnetic resonance images (MRI), which generalizes across MRI sequences and resolutions without retraining. Materials and Methods: We trained our deep learning method, H-synEx, with synthetic images derived from label maps built from ultra-high resolution ex vivo MRI scans, which enables finer-grained manual segmentation when compared with 1mm isometric in vivo images. We validated this retrospective study using 1535 in vivo images from six datasets and six MRI sequences. The quantitative evaluation used the Dice Coefficient (DC) and Average Hausdorff distance (AVD). Statistical analysis compared hypothalamic subregion volumes in controls, Alzheimer's disease (AD), and behavioral variant frontotemporal dementia (bvFTD) subjects using the area under the curve (AUC) and Wilcoxon rank sum test. Results: H-SynEx can segment the hypothalamus across various MRI sequences, encompassing FLAIR sequences with significant slice spacing (5mm). Using hypothalamic volumes on T1w images to distinguish control from AD and bvFTD patients, we observed AUC values of 0.74 and 0.79 respectively. Additionally, AUC=0.66 was found for volume variation on FLAIR scans when comparing control and non-patients. Conclusion: Our results show that H-SynEx successfully leverages information from ultra-high resolution scans to segment in vivo from different MRI sequences such as T1w, T2w, PD, qT1, FA, and FLAIR. We also found that our automated segmentation was able to discriminate controls versus patients on FLAIR images with 5mm spacing. H-SynEx is openly available at https://github.com/liviamarodrigues/hsynex.
Domain-agnostic segmentation of thalamic nuclei from joint structural and diffusion MRI
May 05, 2023Abstract:The human thalamus is a highly connected subcortical grey-matter structure within the brain. It comprises dozens of nuclei with different function and connectivity, which are affected differently by disease. For this reason, there is growing interest in studying the thalamic nuclei in vivo with MRI. Tools are available to segment the thalamus from 1 mm T1 scans, but the contrast of the lateral and internal boundaries is too faint to produce reliable segmentations. Some tools have attempted to incorporate information from diffusion MRI in the segmentation to refine these boundaries, but do not generalise well across diffusion MRI acquisitions. Here we present the first CNN that can segment thalamic nuclei from T1 and diffusion data of any resolution without retraining or fine tuning. Our method builds on a public histological atlas of the thalamic nuclei and silver standard segmentations on high-quality diffusion data obtained with a recent Bayesian adaptive segmentation tool. We combine these with an approximate degradation model for fast domain randomisation during training. Our CNN produces a segmentation at 0.7 mm isotropic resolution, irrespective of the resolution of the input. Moreover, it uses a parsimonious model of the diffusion signal at each voxel (fractional anisotropy and principal eigenvector) that is compatible with virtually any set of directions and b-values, including huge amounts of legacy data. We show results of our proposed method on three heterogeneous datasets acquired on dozens of different scanners. An implementation of the method is publicly available at https://freesurfer.net/fswiki/ThalamicNucleiDTI.
A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology
Jun 22, 2018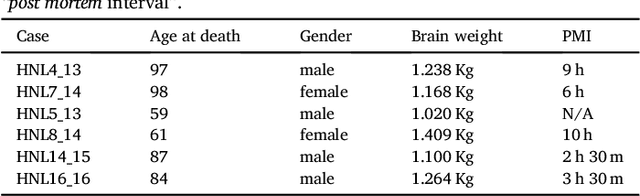
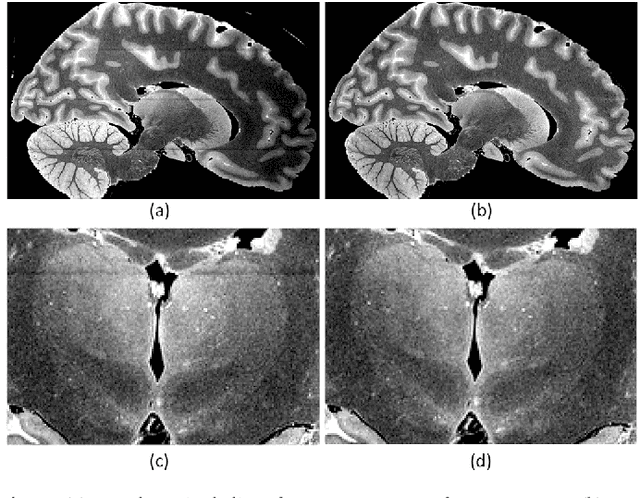
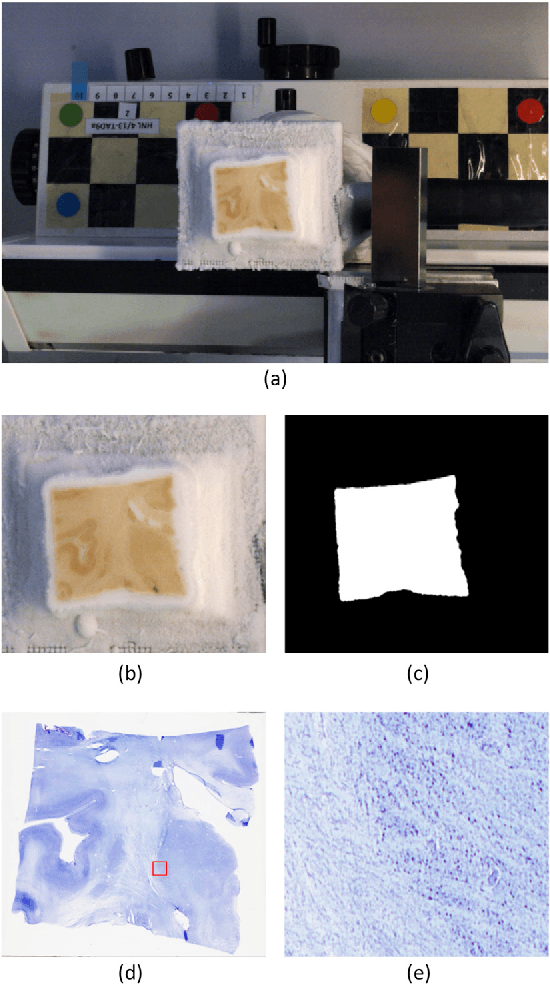
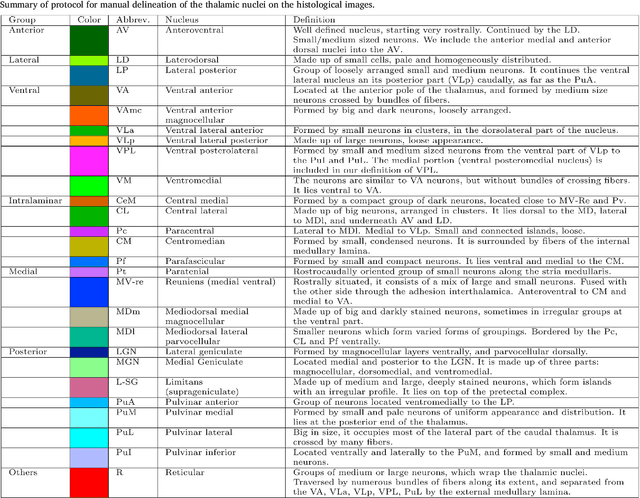
Abstract:The human thalamus is a brain structure that comprises numerous, highly specific nuclei. Since these nuclei are known to have different functions and to be connected to different areas of the cerebral cortex, it is of great interest for the neuroimaging community to study their volume, shape and connectivity in vivo with MRI. In this study, we present a probabilistic atlas of the thalamic nuclei built using ex vivo brain MRI scans and histological data, as well as the application of the atlas to in vivo MRI segmentation. The atlas was built using manual delineation of 26 thalamic nuclei on the serial histology of 12 whole thalami from six autopsy samples, combined with manual segmentations of the whole thalamus and surrounding structures (caudate, putamen, hippocampus, etc.) made on in vivo brain MR data from 39 subjects. The 3D structure of the histological data and corresponding manual segmentations was recovered using the ex vivo MRI as reference frame, and stacks of blockface photographs acquired during the sectioning as intermediate target. The atlas, which was encoded as an adaptive tetrahedral mesh, shows a good agreement with with previous histological studies of the thalamus in terms of volumes of representative nuclei. When applied to segmentation of in vivo scans using Bayesian inference, the atlas shows excellent test-retest reliability, robustness to changes in input MRI contrast, and ability to detect differential thalamic effects in subjects with Alzheimer's disease. The probabilistic atlas and companion segmentation tool are publicly available as part of the neuroimaging package FreeSurfer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge