Vaanathi Sundaresan
Automated quality assessment using appearance-based simulations and hippocampus segmentation on low-field paediatric brain MR images
Oct 08, 2024



Abstract:Understanding the structural growth of paediatric brains is a key step in the identification of various neuro-developmental disorders. However, our knowledge is limited by many factors, including the lack of automated image analysis tools, especially in Low and Middle Income Countries from the lack of high field MR images available. Low-field systems are being increasingly explored in these countries, and, therefore, there is a need to develop automated image analysis tools for these images. In this work, as a preliminary step, we consider two tasks: 1) automated quality assurance and 2) hippocampal segmentation, where we compare multiple approaches. For the automated quality assurance task a DenseNet combined with appearance-based transformations for synthesising artefacts produced the best performance, with a weighted accuracy of 82.3%. For the segmentation task, registration of an average atlas performed the best, with a final Dice score of 0.61. Our results show that although the images can provide understanding of large scale pathologies and gross scale anatomical development, there still remain barriers for their use for more granular analyses.
Class Activation Map-based Weakly supervised Hemorrhage Segmentation using Resnet-LSTM in Non-Contrast Computed Tomography images
Sep 28, 2023



Abstract:In clinical settings, intracranial hemorrhages (ICH) are routinely diagnosed using non-contrast CT (NCCT) for severity assessment. Accurate automated segmentation of ICH lesions is the initial and essential step, immensely useful for such assessment. However, compared to other structural imaging modalities such as MRI, in NCCT images ICH appears with very low contrast and poor SNR. Over recent years, deep learning (DL)-based methods have shown great potential, however, training them requires a huge amount of manually annotated lesion-level labels, with sufficient diversity to capture the characteristics of ICH. In this work, we propose a novel weakly supervised DL method for ICH segmentation on NCCT scans, using image-level binary classification labels, which are less time-consuming and labor-efficient when compared to the manual labeling of individual ICH lesions. Our method initially determines the approximate location of ICH using class activation maps from a classification network, which is trained to learn dependencies across contiguous slices. We further refine the ICH segmentation using pseudo-ICH masks obtained in an unsupervised manner. The method is flexible and uses a computationally light architecture during testing. On evaluating our method on the validation data of the MICCAI 2022 INSTANCE challenge, our method achieves a Dice value of 0.55, comparable with those of existing weakly supervised method (Dice value of 0.47), despite training on a much smaller training data.
Constrained self-supervised method with temporal ensembling for fiber bundle detection on anatomic tracing data
Aug 06, 2022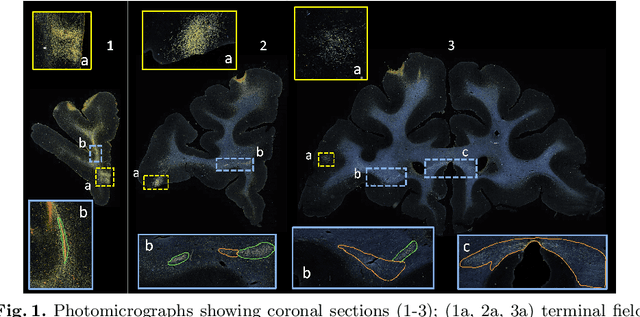
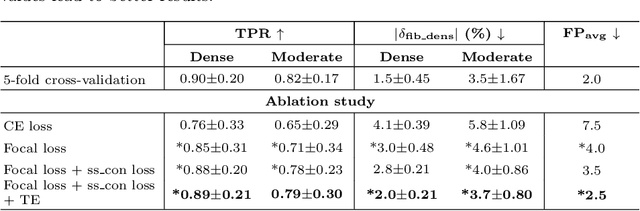
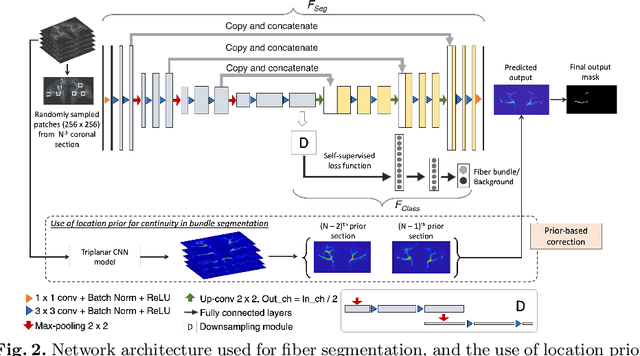
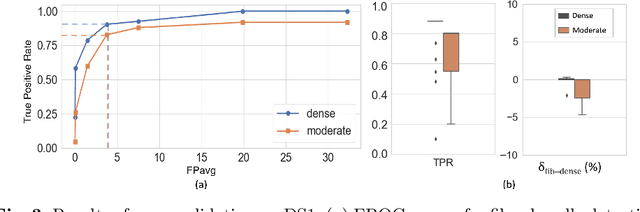
Abstract:Anatomic tracing data provides detailed information on brain circuitry essential for addressing some of the common errors in diffusion MRI tractography. However, automated detection of fiber bundles on tracing data is challenging due to sectioning distortions, presence of noise and artifacts and intensity/contrast variations. In this work, we propose a deep learning method with a self-supervised loss function that takes anatomy-based constraints into account for accurate segmentation of fiber bundles on the tracer sections from macaque brains. Also, given the limited availability of manual labels, we use a semi-supervised training technique for efficiently using unlabeled data to improve the performance, and location constraints for further reduction of false positives. Evaluation of our method on unseen sections from a different macaque yields promising results with a true positive rate of ~0.90. The code for our method is available at https://github.com/v-sundaresan/fiberbundle_seg_tracing.
Challenges for machine learning in clinical translation of big data imaging studies
Jul 07, 2021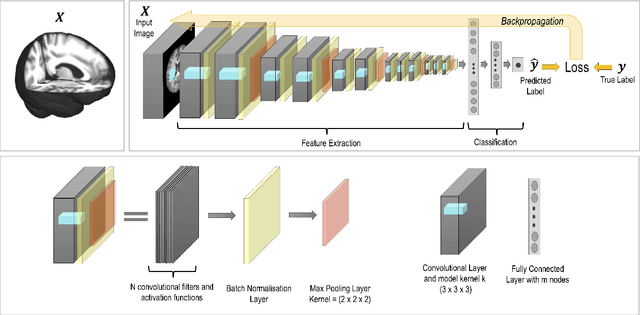
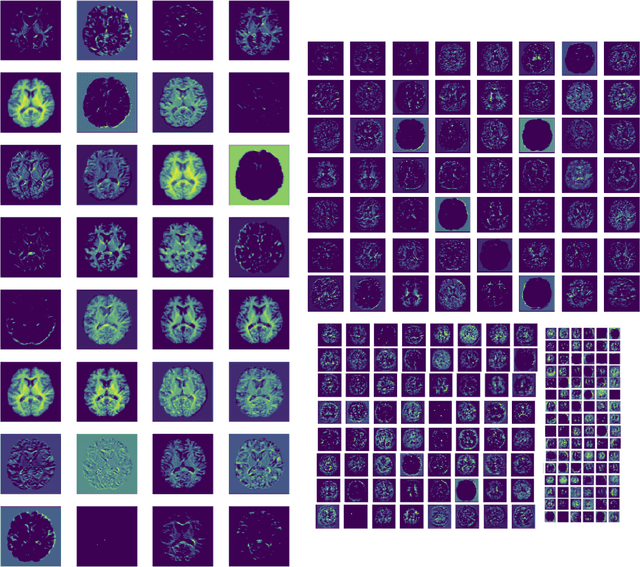
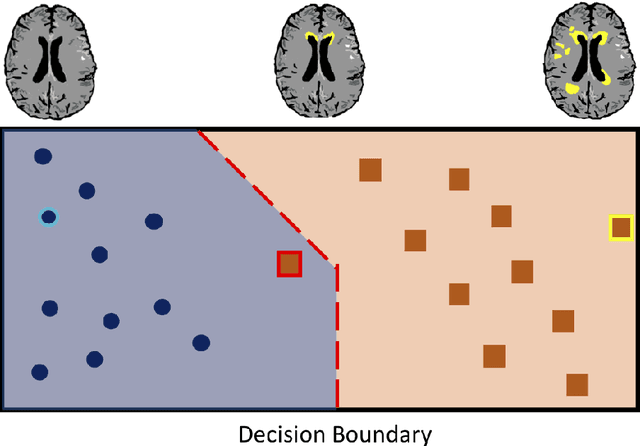
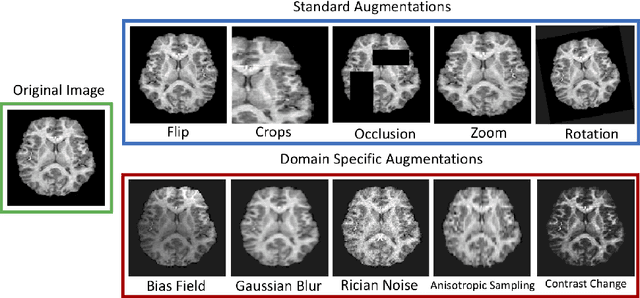
Abstract:The combination of deep learning image analysis methods and large-scale imaging datasets offers many opportunities to imaging neuroscience and epidemiology. However, despite the success of deep learning when applied to many neuroimaging tasks, there remain barriers to the clinical translation of large-scale datasets and processing tools. Here, we explore the main challenges and the approaches that have been explored to overcome them. We focus on issues relating to data availability, interpretability, evaluation and logistical challenges, and discuss the challenges we believe are still to be overcome to enable the full success of big data deep learning approaches to be experienced outside of the research field.
Brain tumour segmentation using a triplanar ensemble of U-Nets
May 24, 2021



Abstract:Gliomas appear with wide variation in their characteristics both in terms of their appearance and location on brain MR images, which makes robust tumour segmentation highly challenging, and leads to high inter-rater variability even in manual segmentations. In this work, we propose a triplanar ensemble network, with an independent tumour core prediction module, for accurate segmentation of these tumours and their sub-regions. On evaluating our method on the MICCAI Brain Tumor Segmentation (BraTS) challenge validation dataset, for tumour sub-regions, we achieved a Dice similarity coefficient of 0.77 for both enhancing tumour (ET) and tumour core (TC). In the case of the whole tumour (WT) region, we achieved a Dice value of 0.89, which is on par with the top-ranking methods from BraTS'17-19. Our method achieved an evaluation score that was the equal 5th highest value (with our method ranking in 10th place) in the BraTS'20 challenge, with mean Dice values of 0.81, 0.89 and 0.84 on ET, WT and TC regions respectively on the BraTS'20 unseen test dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge