Stephen Smith
Coordinated Multi-Neighborhood Learning on a Directed Acyclic Graph
May 24, 2024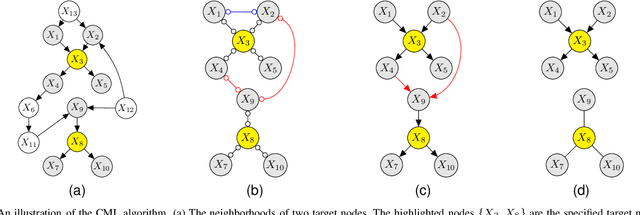
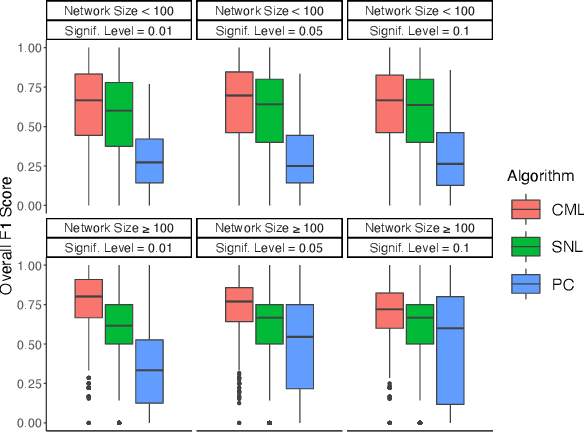
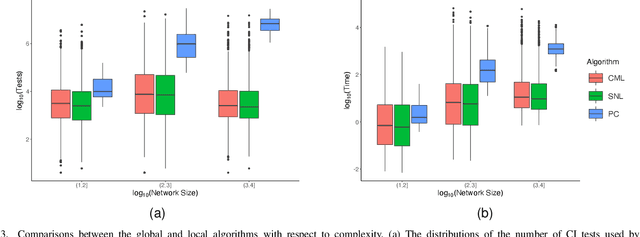
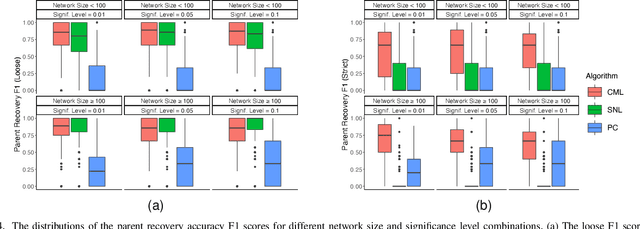
Abstract:Learning the structure of causal directed acyclic graphs (DAGs) is useful in many areas of machine learning and artificial intelligence, with wide applications. However, in the high-dimensional setting, it is challenging to obtain good empirical and theoretical results without strong and often restrictive assumptions. Additionally, it is questionable whether all of the variables purported to be included in the network are observable. It is of interest then to restrict consideration to a subset of the variables for relevant and reliable inferences. In fact, researchers in various disciplines can usually select a set of target nodes in the network for causal discovery. This paper develops a new constraint-based method for estimating the local structure around multiple user-specified target nodes, enabling coordination in structure learning between neighborhoods. Our method facilitates causal discovery without learning the entire DAG structure. We establish consistency results for our algorithm with respect to the local neighborhood structure of the target nodes in the true graph. Experimental results on synthetic and real-world data show that our algorithm is more accurate in learning the neighborhood structures with much less computational cost than standard methods that estimate the entire DAG. An R package implementing our methods may be accessed at https://github.com/stephenvsmith/CML.
Biologically-plausible backpropagation through arbitrary timespans via local neuromodulators
Jun 02, 2022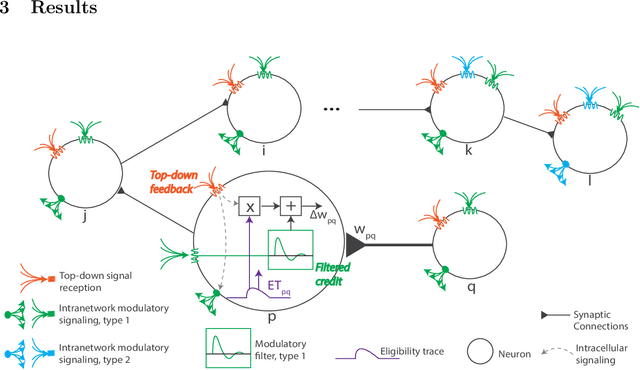
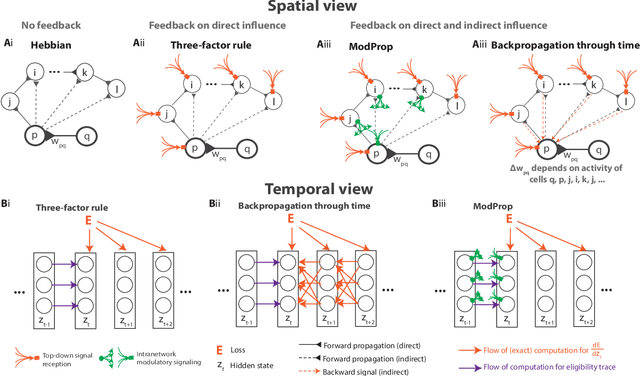
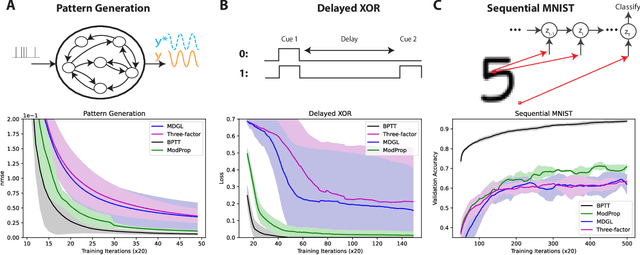
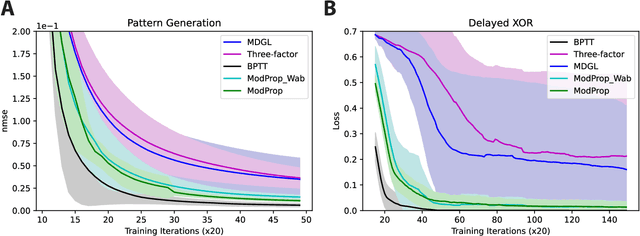
Abstract:The spectacular successes of recurrent neural network models where key parameters are adjusted via backpropagation-based gradient descent have inspired much thought as to how biological neuronal networks might solve the corresponding synaptic credit assignment problem. There is so far little agreement, however, as to how biological networks could implement the necessary backpropagation through time, given widely recognized constraints of biological synaptic network signaling architectures. Here, we propose that extra-synaptic diffusion of local neuromodulators such as neuropeptides may afford an effective mode of backpropagation lying within the bounds of biological plausibility. Going beyond existing temporal truncation-based gradient approximations, our approximate gradient-based update rule, ModProp, propagates credit information through arbitrary time steps. ModProp suggests that modulatory signals can act on receiving cells by convolving their eligibility traces via causal, time-invariant and synapse-type-specific filter taps. Our mathematical analysis of ModProp learning, together with simulation results on benchmark temporal tasks, demonstrate the advantage of ModProp over existing biologically-plausible temporal credit assignment rules. These results suggest a potential neuronal mechanism for signaling credit information related to recurrent interactions over a longer time horizon. Finally, we derive an in-silico implementation of ModProp that could serve as a low-complexity and causal alternative to backpropagation through time.
Challenges for machine learning in clinical translation of big data imaging studies
Jul 07, 2021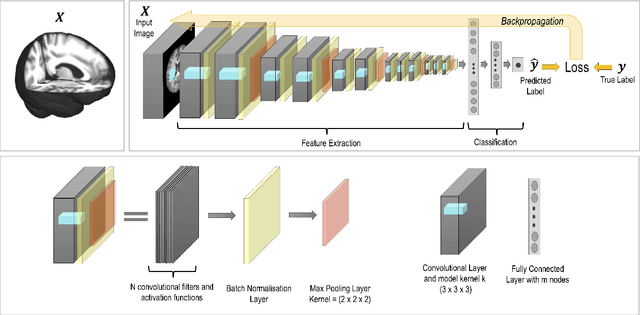
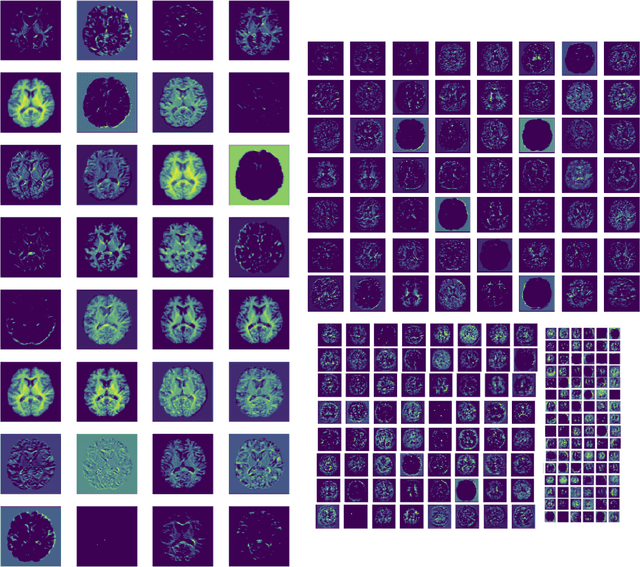
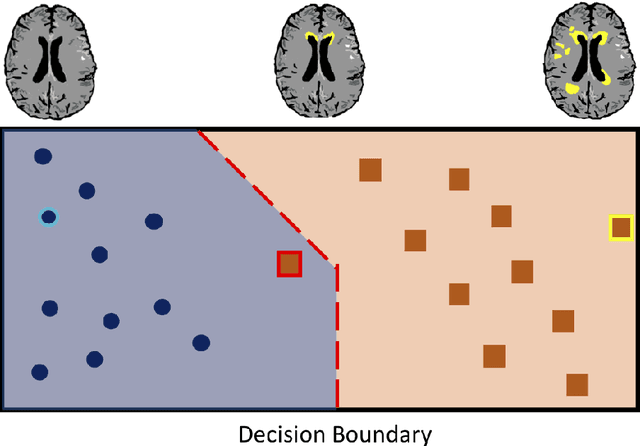
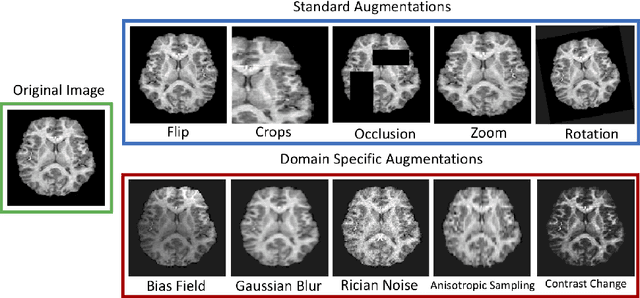
Abstract:The combination of deep learning image analysis methods and large-scale imaging datasets offers many opportunities to imaging neuroscience and epidemiology. However, despite the success of deep learning when applied to many neuroimaging tasks, there remain barriers to the clinical translation of large-scale datasets and processing tools. Here, we explore the main challenges and the approaches that have been explored to overcome them. We focus on issues relating to data availability, interpretability, evaluation and logistical challenges, and discuss the challenges we believe are still to be overcome to enable the full success of big data deep learning approaches to be experienced outside of the research field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge