Stefan Mihalas
Slow Transition to Low-Dimensional Chaos in Heavy-Tailed Recurrent Neural Networks
May 14, 2025Abstract:Growing evidence suggests that synaptic weights in the brain follow heavy-tailed distributions, yet most theoretical analyses of recurrent neural networks (RNNs) assume Gaussian connectivity. We systematically study the activity of RNNs with random weights drawn from biologically plausible L\'evy alpha-stable distributions. While mean-field theory for the infinite system predicts that the quiescent state is always unstable -- implying ubiquitous chaos -- our finite-size analysis reveals a sharp transition between quiescent and chaotic dynamics. We theoretically predict the gain at which the system transitions from quiescent to chaotic dynamics, and validate it through simulations. Compared to Gaussian networks, heavy-tailed RNNs exhibit a broader parameter regime near the edge of chaos, namely a slow transition to chaos. However, this robustness comes with a tradeoff: heavier tails reduce the Lyapunov dimension of the attractor, indicating lower effective dimensionality. Our results reveal a biologically aligned tradeoff between the robustness of dynamics near the edge of chaos and the richness of high-dimensional neural activity. By analytically characterizing the transition point in finite-size networks -- where mean-field theory breaks down -- we provide a tractable framework for understanding dynamics in realistically sized, heavy-tailed neural circuits.
A Fokker-Planck-Based Loss Function that Bridges Dynamics with Density Estimation
Feb 24, 2025Abstract:We have derived a novel loss function from the Fokker-Planck equation that links dynamical system models with their probability density functions, demonstrating its utility in model identification and density estimation. In the first application, we show that this loss function can enable the extraction of dynamical parameters from non-temporal datasets, including timestamp-free measurements from steady non-equilibrium systems such as noisy Lorenz systems and gene regulatory networks. In the second application, when coupled with a density estimator, this loss facilitates density estimation when the dynamic equations are known. For density estimation, we propose a density estimator that integrates a Gaussian Mixture Model with a normalizing flow model. It simultaneously estimates normalized density, energy, and score functions from both empirical data and dynamics. It is compatible with a variety of data-based training methodologies, including maximum likelihood and score matching. It features a latent space akin to a modern Hopfield network, where the inherent Hopfield energy effectively assigns low densities to sparsely populated data regions, addressing common challenges in neural density estimators. Additionally, this Hopfield-like energy enables direct and rapid data manipulation through the Concave-Convex Procedure (CCCP) rule, facilitating tasks such as denoising and clustering. Our work demonstrates a principled framework for leveraging the complex interdependencies between dynamics and density estimation, as illustrated through synthetic examples that clarify the underlying theoretical intuitions.
Hierarchy of chaotic dynamics in random modular networks
Oct 08, 2024Abstract:We introduce a model of randomly connected neural populations and study its dynamics by means of the dynamical mean-field theory and simulations. Our analysis uncovers a rich phase diagram, featuring high- and low-dimensional chaotic phases, separated by a crossover region characterized by low values of the maximal Lyapunov exponent and participation ratio dimension, but with high and rapidly changing values of the Lyapunov dimension. Counterintuitively, chaos can be attenuated by either adding noise to strongly modular connectivity or by introducing modularity into random connectivity. Extending the model to include a multilevel, hierarchical connectivity reveals that a loose balance between activities across levels drives the system towards the edge of chaos.
How connectivity structure shapes rich and lazy learning in neural circuits
Oct 12, 2023Abstract:In theoretical neuroscience, recent work leverages deep learning tools to explore how some network attributes critically influence its learning dynamics. Notably, initial weight distributions with small (resp. large) variance may yield a rich (resp. lazy) regime, where significant (resp. minor) changes to network states and representation are observed over the course of learning. However, in biology, neural circuit connectivity generally has a low-rank structure and therefore differs markedly from the random initializations generally used for these studies. As such, here we investigate how the structure of the initial weights, in particular their effective rank, influences the network learning regime. Through both empirical and theoretical analyses, we discover that high-rank initializations typically yield smaller network changes indicative of lazier learning, a finding we also confirm with experimentally-driven initial connectivity in recurrent neural networks. Conversely, low-rank initialization biases learning towards richer learning. Importantly, however, as an exception to this rule, we find lazier learning can still occur with a low-rank initialization that aligns with task and data statistics. Our research highlights the pivotal role of initial weight structures in shaping learning regimes, with implications for metabolic costs of plasticity and risks of catastrophic forgetting.
Biologically-plausible backpropagation through arbitrary timespans via local neuromodulators
Jun 02, 2022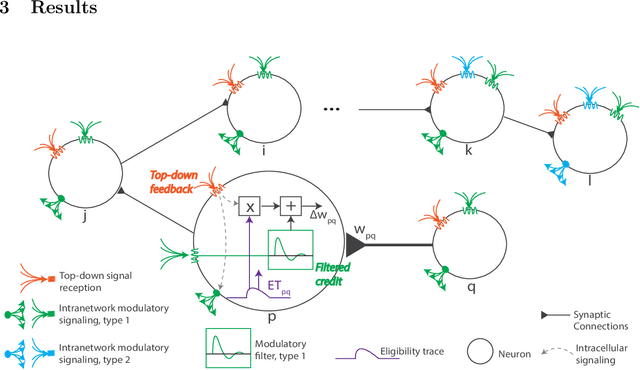
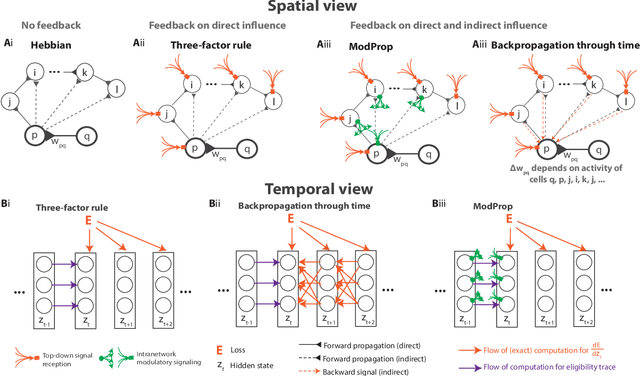
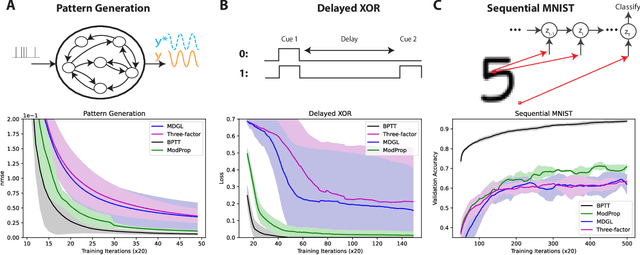
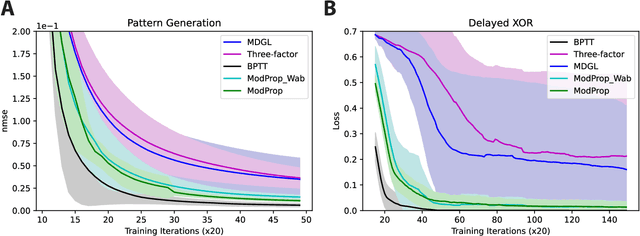
Abstract:The spectacular successes of recurrent neural network models where key parameters are adjusted via backpropagation-based gradient descent have inspired much thought as to how biological neuronal networks might solve the corresponding synaptic credit assignment problem. There is so far little agreement, however, as to how biological networks could implement the necessary backpropagation through time, given widely recognized constraints of biological synaptic network signaling architectures. Here, we propose that extra-synaptic diffusion of local neuromodulators such as neuropeptides may afford an effective mode of backpropagation lying within the bounds of biological plausibility. Going beyond existing temporal truncation-based gradient approximations, our approximate gradient-based update rule, ModProp, propagates credit information through arbitrary time steps. ModProp suggests that modulatory signals can act on receiving cells by convolving their eligibility traces via causal, time-invariant and synapse-type-specific filter taps. Our mathematical analysis of ModProp learning, together with simulation results on benchmark temporal tasks, demonstrate the advantage of ModProp over existing biologically-plausible temporal credit assignment rules. These results suggest a potential neuronal mechanism for signaling credit information related to recurrent interactions over a longer time horizon. Finally, we derive an in-silico implementation of ModProp that could serve as a low-complexity and causal alternative to backpropagation through time.
Convolutional neural networks with extra-classical receptive fields
Oct 27, 2018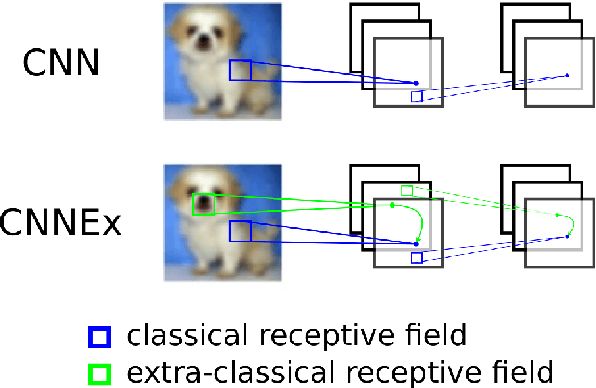
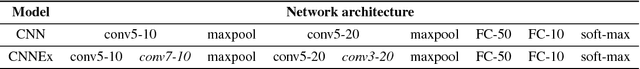
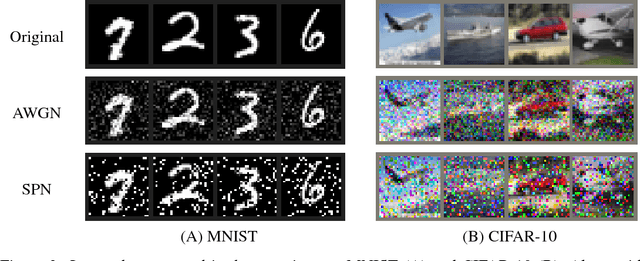
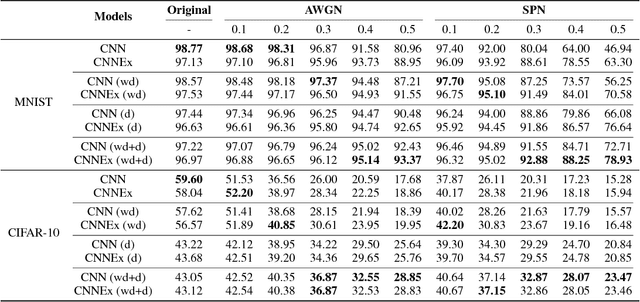
Abstract:Convolutional neural networks (CNNs) have had great success in many real-world applications and have also been used to model visual processing in the brain. However, these networks are quite brittle - small changes in the input image can dramatically change a network's output prediction. In contrast to what is known from biology, these networks largely rely on feedforward connections, ignoring the influence of recurrent connections. They also focus on supervised rather than unsupervised learning. To address these issues, we combine traditional supervised learning via backpropagation with a specialized unsupervised learning rule to learn lateral connections between neurons within a convolutional neural network. These connections have been shown to optimally integrate information from the surround, generating extra-classical receptive fields for the neurons in our new proposed model (CNNEx). Models with optimal lateral connections are more robust to noise and achieve better performance on noisy versions of the MNIST and CIFAR-10 datasets. Resistance to noise can be further improved by combining our model with additional regularization techniques such as dropout and weight decay. Although the image statistics of MNIST and CIFAR-10 differ greatly, the same unsupervised learning rule generalized to both datasets. Our results demonstrate the potential usefulness of combining supervised and unsupervised learning techniques and suggest that the integration of lateral connections into convolutional neural networks is an important area of future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge