Mark Jenkinson
Australian Institute for Machine Learning, South Australian Health and Medical Research Institute
Validation of a CT-brain analysis tool for measuring global cortical atrophy in older patient cohorts
Sep 08, 2025Abstract:Quantification of brain atrophy currently requires visual rating scales which are time consuming and automated brain image analysis is warranted. We validated our automated deep learning (DL) tool measuring the Global Cerebral Atrophy (GCA) score against trained human raters, and associations with age and cognitive impairment, in representative older (>65 years) patients. CT-brain scans were obtained from patients in acute medicine (ORCHARD-EPR), acute stroke (OCS studies) and a legacy sample. Scans were divided in a 60/20/20 ratio for training, optimisation and testing. CT-images were assessed by two trained raters (rater-1=864 scans, rater-2=20 scans). Agreement between DL tool-predicted GCA scores (range 0-39) and the visual ratings was evaluated using mean absolute error (MAE) and Cohen's weighted kappa. Among 864 scans (ORCHARD-EPR=578, OCS=200, legacy scans=86), MAE between the DL tool and rater-1 GCA scores was 3.2 overall, 3.1 for ORCHARD-EPR, 3.3 for OCS and 2.6 for the legacy scans and half had DL-predicted GCA error between -2 and 2. Inter-rater agreement was Kappa=0.45 between the DL-tool and rater-1, and 0.41 between the tool and rater- 2 whereas it was lower at 0.28 for rater-1 and rater-2. There was no difference in GCA scores from the DL-tool and the two raters (one-way ANOVA, p=0.35) or in mean GCA scores between the DL-tool and rater-1 (paired t-test, t=-0.43, p=0.66), the tool and rater-2 (t=1.35, p=0.18) or between rater-1 and rater-2 (t=0.99, p=0.32). DL-tool GCA scores correlated with age and cognitive scores (both p<0.001). Our DL CT-brain analysis tool measured GCA score accurately and without user input in real-world scans acquired from older patients. Our tool will enable extraction of standardised quantitative measures of atrophy at scale for use in health data research and will act as proof-of-concept towards a point-of-care clinically approved tool.
SFHarmony: Source Free Domain Adaptation for Distributed Neuroimaging Analysis
Mar 28, 2023



Abstract:To represent the biological variability of clinical neuroimaging populations, it is vital to be able to combine data across scanners and studies. However, different MRI scanners produce images with different characteristics, resulting in a domain shift known as the `harmonisation problem'. Additionally, neuroimaging data is inherently personal in nature, leading to data privacy concerns when sharing the data. To overcome these barriers, we propose an Unsupervised Source-Free Domain Adaptation (SFDA) method, SFHarmony. Through modelling the imaging features as a Gaussian Mixture Model and minimising an adapted Bhattacharyya distance between the source and target features, we can create a model that performs well for the target data whilst having a shared feature representation across the data domains, without needing access to the source data for adaptation or target labels. We demonstrate the performance of our method on simulated and real domain shifts, showing that the approach is applicable to classification, segmentation and regression tasks, requiring no changes to the algorithm. Our method outperforms existing SFDA approaches across a range of realistic data scenarios, demonstrating the potential utility of our approach for MRI harmonisation and general SFDA problems. Our code is available at \url{https://github.com/nkdinsdale/SFHarmony}.
Segmentation method for cerebral blood vessels from MRA using hysteresis
Mar 09, 2023Abstract:Segmentation of cerebral blood vessels from Magnetic Resonance Imaging (MRI) is an open problem that could be solved with deep learning (DL). However, annotated data for training is often scarce. Due to the absence of open-source tools, we aim to develop a classical segmentation method that generates vessel ground truth from Magnetic Resonance Angiography for DL training of segmentation across a variety of modalities. The method combines size-specific Hessian filters, hysteresis thresholding and connected component correction. The optimal choice of processing steps was evaluated with a blinded scoring by a clinician using 24 3D images. The results show that all method steps are necessary to produce the highest (14.2/15) vessel segmentation quality score. Omitting the connected component correction caused the largest quality loss. The method, which is available on GitHub, can be used to train DL models for vessel segmentation.
FedHarmony: Unlearning Scanner Bias with Distributed Data
May 31, 2022



Abstract:The ability to combine data across scanners and studies is vital for neuroimaging, to increase both statistical power and the representation of biological variability. However, combining datasets across sites leads to two challenges: first, an increase in undesirable non-biological variance due to scanner and acquisition differences - the harmonisation problem - and second, data privacy concerns due to the inherently personal nature of medical imaging data, meaning that sharing them across sites may risk violation of privacy laws. To overcome these restrictions, we propose FedHarmony: a harmonisation framework operating in the federated learning paradigm. We show that to remove the scanner-specific effects, we only need to share the mean and standard deviation of the learned features, helping to protect individual subjects' privacy. We demonstrate our approach across a range of realistic data scenarios, using real multi-site data from the ABIDE dataset, thus showing the potential utility of our method for MRI harmonisation across studies. Our code is available at https://github.com/nkdinsdale/FedHarmony.
How certain are your uncertainties?
Mar 01, 2022


Abstract:Having a measure of uncertainty in the output of a deep learning method is useful in several ways, such as in assisting with interpretation of the outputs, helping build confidence with end users, and for improving the training and performance of the networks. Therefore, several different methods have been proposed to capture various types of uncertainty, including epistemic (relating to the model used) and aleatoric (relating to the data) sources, with the most commonly used methods for estimating these being test-time dropout for epistemic uncertainty and test-time augmentation for aleatoric uncertainty. However, these methods are parameterised (e.g. amount of dropout or type and level of augmentation) and so there is a whole range of possible uncertainties that could be calculated, even with a fixed network and dataset. This work investigates the stability of these uncertainty measurements, in terms of both magnitude and spatial pattern. In experiments using the well characterised BraTS challenge, we demonstrate substantial variability in the magnitude and spatial pattern of these uncertainties, and discuss the implications for interpretability, repeatability and confidence in results.
Mutual information neural estimation for unsupervised multi-modal registration of brain images
Jan 25, 2022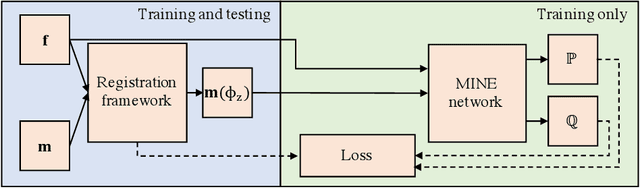
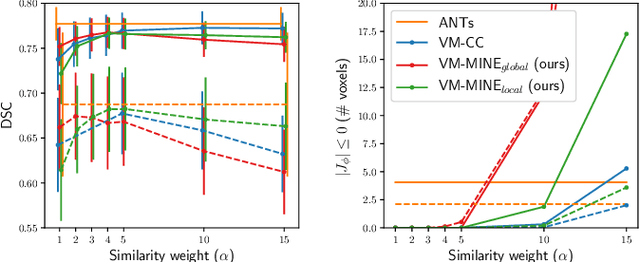
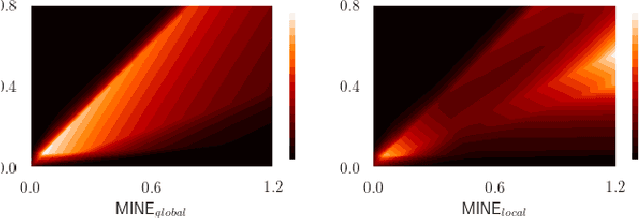

Abstract:Many applications in image-guided surgery and therapy require fast and reliable non-linear, multi-modal image registration. Recently proposed unsupervised deep learning-based registration methods have demonstrated superior performance compared to iterative methods in just a fraction of the time. Most of the learning-based methods have focused on mono-modal image registration. The extension to multi-modal registration depends on the use of an appropriate similarity function, such as the mutual information (MI). We propose guiding the training of a deep learning-based registration method with MI estimation between an image-pair in an end-to-end trainable network. Our results show that a small, 2-layer network produces competitive results in both mono- and multimodal registration, with sub-second run-times. Comparisons to both iterative and deep learning-based methods show that our MI-based method produces topologically and qualitatively superior results with an extremely low rate of non-diffeomorphic transformations. Real-time clinical application will benefit from a better visual matching of anatomical structures and less registration failures/outliers.
TEDS-Net: Enforcing Diffeomorphisms in Spatial Transformers to Guarantee Topology Preservation in Segmentations
Jul 28, 2021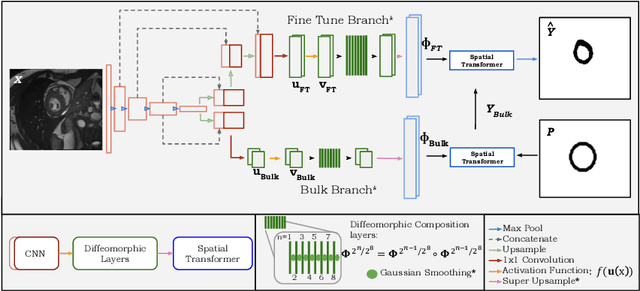
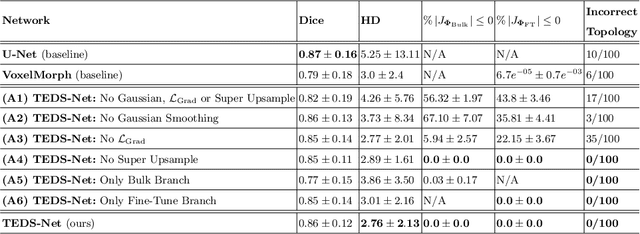


Abstract:Accurate topology is key when performing meaningful anatomical segmentations, however, it is often overlooked in traditional deep learning methods. In this work we propose TEDS-Net: a novel segmentation method that guarantees accurate topology. Our method is built upon a continuous diffeomorphic framework, which enforces topology preservation. However, in practice, diffeomorphic fields are represented using a finite number of parameters and sampled using methods such as linear interpolation, violating the theoretical guarantees. We therefore introduce additional modifications to more strictly enforce it. Our network learns how to warp a binary prior, with the desired topological characteristics, to complete the segmentation task. We tested our method on myocardium segmentation from an open-source 2D heart dataset. TEDS-Net preserved topology in 100% of the cases, compared to 90% from the U-Net, without sacrificing on Hausdorff Distance or Dice performance. Code will be made available at: www.github.com/mwyburd/TEDS-Net
Challenges for machine learning in clinical translation of big data imaging studies
Jul 07, 2021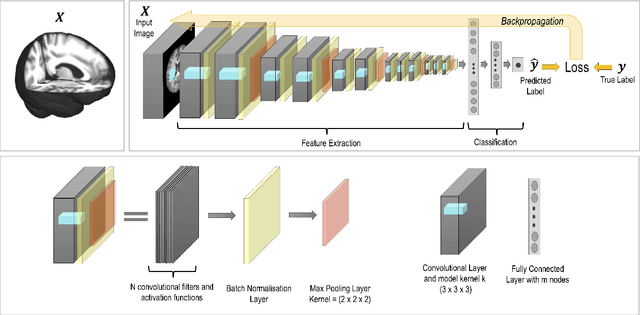
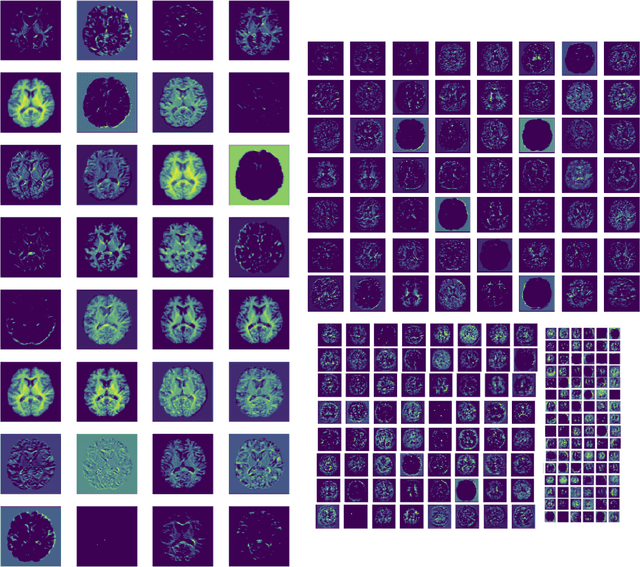
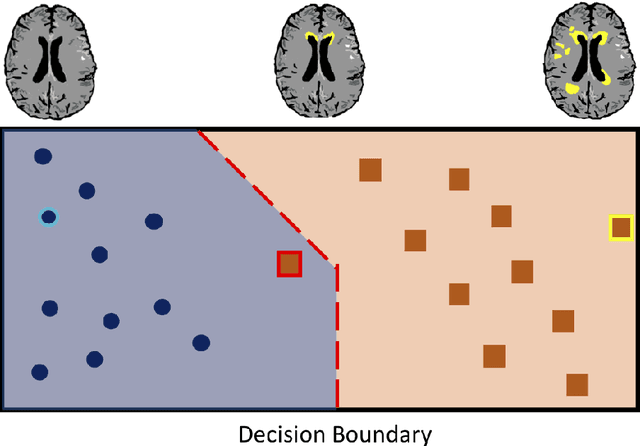
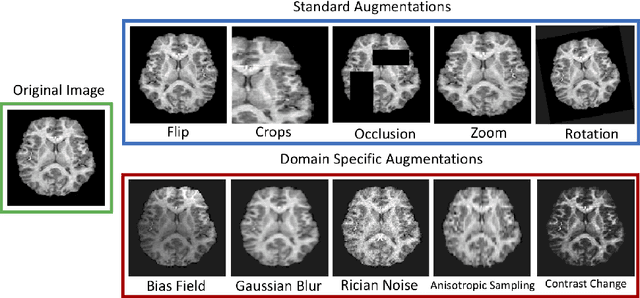
Abstract:The combination of deep learning image analysis methods and large-scale imaging datasets offers many opportunities to imaging neuroscience and epidemiology. However, despite the success of deep learning when applied to many neuroimaging tasks, there remain barriers to the clinical translation of large-scale datasets and processing tools. Here, we explore the main challenges and the approaches that have been explored to overcome them. We focus on issues relating to data availability, interpretability, evaluation and logistical challenges, and discuss the challenges we believe are still to be overcome to enable the full success of big data deep learning approaches to be experienced outside of the research field.
Self-supervised Lesion Change Detection and Localisation in Longitudinal Multiple Sclerosis Brain Imaging
Jun 02, 2021



Abstract:Longitudinal imaging forms an essential component in the management and follow-up of many medical conditions. The presence of lesion changes on serial imaging can have significant impact on clinical decision making, highlighting the important role for automated change detection. Lesion changes can represent anomalies in serial imaging, which implies a limited availability of annotations and a wide variety of possible changes that need to be considered. Hence, we introduce a new unsupervised anomaly detection and localisation method trained exclusively with serial images that do not contain any lesion changes. Our training automatically synthesises lesion changes in serial images, introducing detection and localisation pseudo-labels that are used to self-supervise the training of our model. Given the rarity of these lesion changes in the synthesised images, we train the model with the imbalance robust focal Tversky loss. When compared to supervised models trained on different datasets, our method shows competitive performance in the detection and localisation of new demyelinating lesions on longitudinal magnetic resonance imaging in multiple sclerosis patients. Code for the models will be made available on GitHub.
Brain tumour segmentation using a triplanar ensemble of U-Nets
May 24, 2021



Abstract:Gliomas appear with wide variation in their characteristics both in terms of their appearance and location on brain MR images, which makes robust tumour segmentation highly challenging, and leads to high inter-rater variability even in manual segmentations. In this work, we propose a triplanar ensemble network, with an independent tumour core prediction module, for accurate segmentation of these tumours and their sub-regions. On evaluating our method on the MICCAI Brain Tumor Segmentation (BraTS) challenge validation dataset, for tumour sub-regions, we achieved a Dice similarity coefficient of 0.77 for both enhancing tumour (ET) and tumour core (TC). In the case of the whole tumour (WT) region, we achieved a Dice value of 0.89, which is on par with the top-ranking methods from BraTS'17-19. Our method achieved an evaluation score that was the equal 5th highest value (with our method ranking in 10th place) in the BraTS'20 challenge, with mean Dice values of 0.81, 0.89 and 0.84 on ET, WT and TC regions respectively on the BraTS'20 unseen test dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge