Ana I. L. Namburete
UltraGauss: Ultrafast Gaussian Reconstruction of 3D Ultrasound Volumes
May 08, 2025Abstract:Ultrasound imaging is widely used due to its safety, affordability, and real-time capabilities, but its 2D interpretation is highly operator-dependent, leading to variability and increased cognitive demand. 2D-to-3D reconstruction mitigates these challenges by providing standardized volumetric views, yet existing methods are often computationally expensive, memory-intensive, or incompatible with ultrasound physics. We introduce UltraGauss: the first ultrasound-specific Gaussian Splatting framework, extending view synthesis techniques to ultrasound wave propagation. Unlike conventional perspective-based splatting, UltraGauss models probe-plane intersections in 3D, aligning with acoustic image formation. We derive an efficient rasterization boundary formulation for GPU parallelization and introduce a numerically stable covariance parametrization, improving computational efficiency and reconstruction accuracy. On real clinical ultrasound data, UltraGauss achieves state-of-the-art reconstructions in 5 minutes, and reaching 0.99 SSIM within 20 minutes on a single GPU. A survey of expert clinicians confirms UltraGauss' reconstructions are the most realistic among competing methods. Our CUDA implementation will be released upon publication.
Exploring Test Time Adaptation for Subcortical Segmentation of the Fetal Brain in 3D Ultrasound
Feb 12, 2025Abstract:Monitoring the growth of subcortical regions of the fetal brain in ultrasound (US) images can help identify the presence of abnormal development. Manually segmenting these regions is a challenging task, but recent work has shown that it can be automated using deep learning. However, applying pretrained models to unseen freehand US volumes often leads to a degradation of performance due to the vast differences in acquisition and alignment. In this work, we first demonstrate that test time adaptation (TTA) can be used to improve model performance in the presence of both real and simulated domain shifts. We further propose a novel TTA method by incorporating a normative atlas as a prior for anatomy. In the presence of various types of domain shifts, we benchmark the performance of different TTA methods and demonstrate the improvements brought by our proposed approach, which may further facilitate automated monitoring of fetal brain development. Our code is available at https://github.com/joshuaomolegan/TTA-for-3D-Fetal-Subcortical-Segmentation.
RapidVol: Rapid Reconstruction of 3D Ultrasound Volumes from Sensorless 2D Scans
Apr 16, 2024



Abstract:Two-dimensional (2D) freehand ultrasonography is one of the most commonly used medical imaging modalities, particularly in obstetrics and gynaecology. However, it only captures 2D cross-sectional views of inherently 3D anatomies, losing valuable contextual information. As an alternative to requiring costly and complex 3D ultrasound scanners, 3D volumes can be constructed from 2D scans using machine learning. However this usually requires long computational time. Here, we propose RapidVol: a neural representation framework to speed up slice-to-volume ultrasound reconstruction. We use tensor-rank decomposition, to decompose the typical 3D volume into sets of tri-planes, and store those instead, as well as a small neural network. A set of 2D ultrasound scans, with their ground truth (or estimated) 3D position and orientation (pose) is all that is required to form a complete 3D reconstruction. Reconstructions are formed from real fetal brain scans, and then evaluated by requesting novel cross-sectional views. When compared to prior approaches based on fully implicit representation (e.g. neural radiance fields), our method is over 3x quicker, 46% more accurate, and if given inaccurate poses is more robust. Further speed-up is also possible by reconstructing from a structural prior rather than from scratch.
Prototype Learning for Explainable Regression
Jun 16, 2023Abstract:The lack of explainability limits the adoption of deep learning models in clinical practice. While methods exist to improve the understanding of such models, these are mainly saliency-based and developed for classification, despite many important tasks in medical imaging being continuous regression problems. Therefore, in this work, we present ExPeRT: an explainable prototype-based model specifically designed for regression tasks. Our proposed model makes a sample prediction from the distances to a set of learned prototypes in latent space, using a weighted mean of prototype labels. The distances in latent space are regularized to be relative to label differences, and each of the prototypes can be visualized as a sample from the training set. The image-level distances are further constructed from patch-level distances, in which the patches of both images are structurally matched using optimal transport. We demonstrate our proposed model on the task of brain age prediction on two image datasets: adult MR and fetal ultrasound. Our approach achieved state-of-the-art prediction performance while providing insight in the model's reasoning process.
Adaptive 3D Localization of 2D Freehand Ultrasound Brain Images
Sep 12, 2022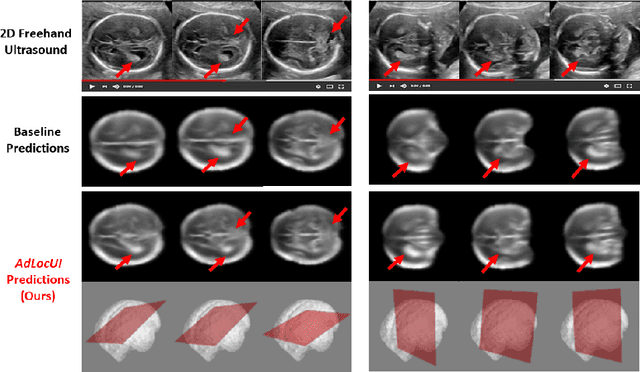
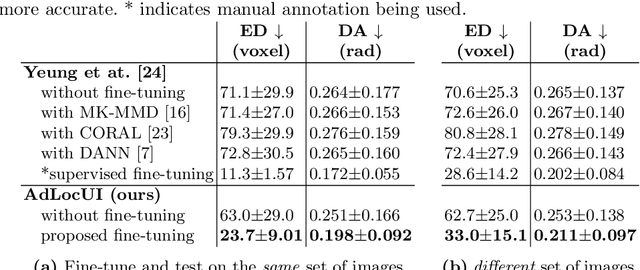
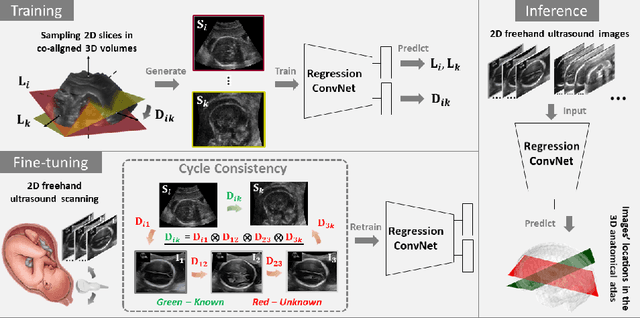
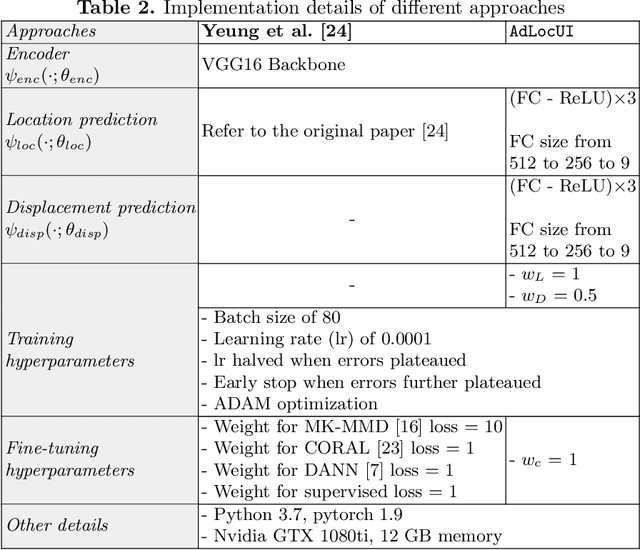
Abstract:Two-dimensional (2D) freehand ultrasound is the mainstay in prenatal care and fetal growth monitoring. The task of matching corresponding cross-sectional planes in the 3D anatomy for a given 2D ultrasound brain scan is essential in freehand scanning, but challenging. We propose AdLocUI, a framework that Adaptively Localizes 2D Ultrasound Images in the 3D anatomical atlas without using any external tracking sensor.. We first train a convolutional neural network with 2D slices sampled from co-aligned 3D ultrasound volumes to predict their locations in the 3D anatomical atlas. Next, we fine-tune it with 2D freehand ultrasound images using a novel unsupervised cycle consistency, which utilizes the fact that the overall displacement of a sequence of images in the 3D anatomical atlas is equal to the displacement from the first image to the last in that sequence. We demonstrate that AdLocUI can adapt to three different ultrasound datasets, acquired with different machines and protocols, and achieves significantly better localization accuracy than the baselines. AdLocUI can be used for sensorless 2D freehand ultrasound guidance by the bedside. The source code is available at https://github.com/pakheiyeung/AdLocUI.
INSightR-Net: Interpretable Neural Network for Regression using Similarity-based Comparisons to Prototypical Examples
Jul 31, 2022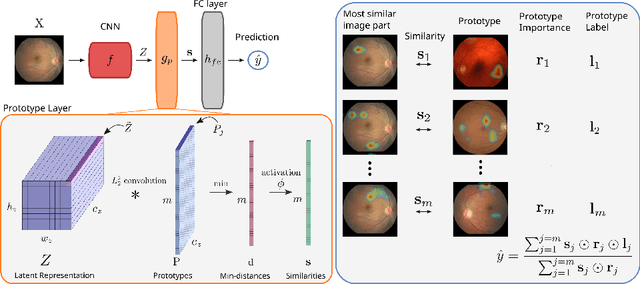
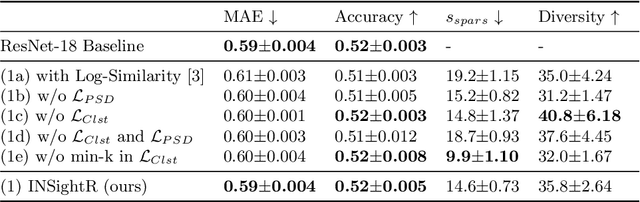
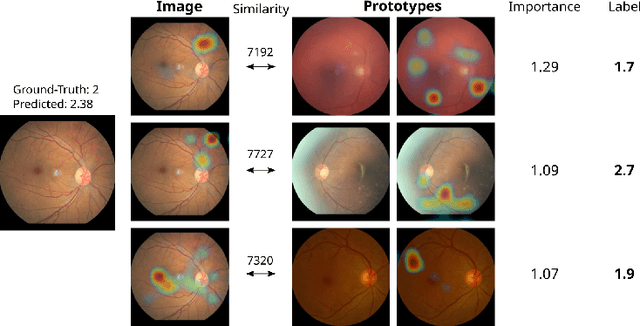
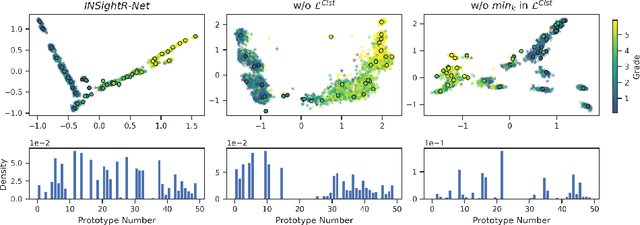
Abstract:Convolutional neural networks (CNNs) have shown exceptional performance for a range of medical imaging tasks. However, conventional CNNs are not able to explain their reasoning process, therefore limiting their adoption in clinical practice. In this work, we propose an inherently interpretable CNN for regression using similarity-based comparisons (INSightR-Net) and demonstrate our methods on the task of diabetic retinopathy grading. A prototype layer incorporated into the architecture enables visualization of the areas in the image that are most similar to learned prototypes. The final prediction is then intuitively modeled as a mean of prototype labels, weighted by the similarities. We achieved competitive prediction performance with our INSightR-Net compared to a ResNet baseline, showing that it is not necessary to compromise performance for interpretability. Furthermore, we quantified the quality of our explanations using sparsity and diversity, two concepts considered important for a good explanation, and demonstrated the effect of several parameters on the latent space embeddings.
ImplicitVol: Sensorless 3D Ultrasound Reconstruction with Deep Implicit Representation
Sep 24, 2021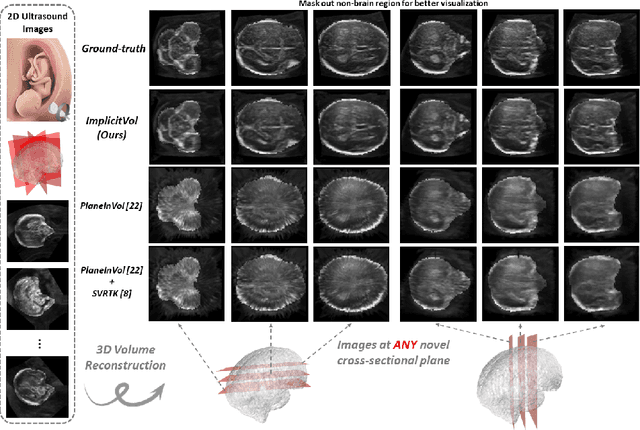
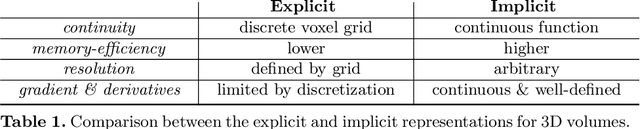
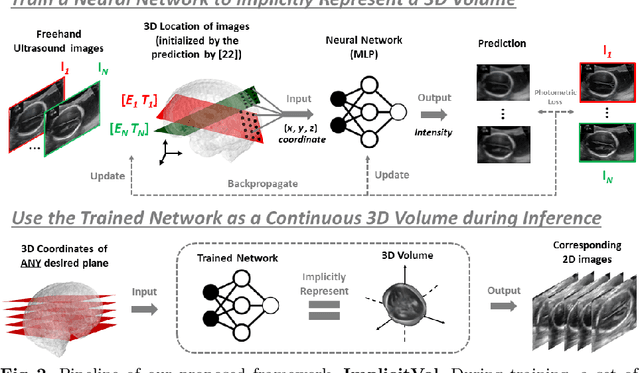
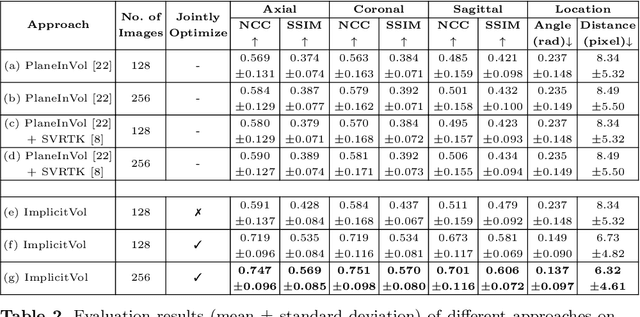
Abstract:The objective of this work is to achieve sensorless reconstruction of a 3D volume from a set of 2D freehand ultrasound images with deep implicit representation. In contrast to the conventional way that represents a 3D volume as a discrete voxel grid, we do so by parameterizing it as the zero level-set of a continuous function, i.e. implicitly representing the 3D volume as a mapping from the spatial coordinates to the corresponding intensity values. Our proposed model, termed as ImplicitVol, takes a set of 2D scans and their estimated locations in 3D as input, jointly re?fing the estimated 3D locations and learning a full reconstruction of the 3D volume. When testing on real 2D ultrasound images, novel cross-sectional views that are sampled from ImplicitVol show significantly better visual quality than those sampled from existing reconstruction approaches, outperforming them by over 30% (NCC and SSIM), between the output and ground-truth on the 3D volume testing data. The code will be made publicly available.
TEDS-Net: Enforcing Diffeomorphisms in Spatial Transformers to Guarantee Topology Preservation in Segmentations
Jul 28, 2021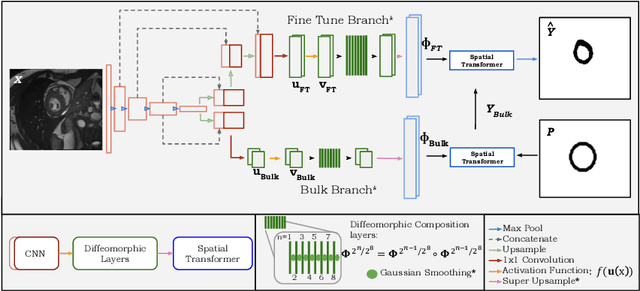
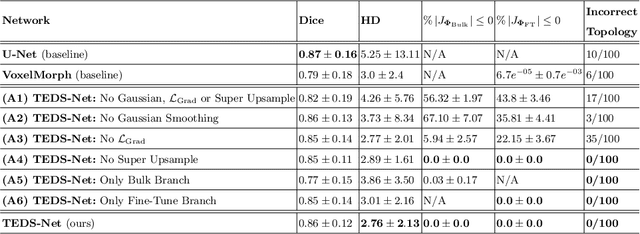


Abstract:Accurate topology is key when performing meaningful anatomical segmentations, however, it is often overlooked in traditional deep learning methods. In this work we propose TEDS-Net: a novel segmentation method that guarantees accurate topology. Our method is built upon a continuous diffeomorphic framework, which enforces topology preservation. However, in practice, diffeomorphic fields are represented using a finite number of parameters and sampled using methods such as linear interpolation, violating the theoretical guarantees. We therefore introduce additional modifications to more strictly enforce it. Our network learns how to warp a binary prior, with the desired topological characteristics, to complete the segmentation task. We tested our method on myocardium segmentation from an open-source 2D heart dataset. TEDS-Net preserved topology in 100% of the cases, compared to 90% from the U-Net, without sacrificing on Hausdorff Distance or Dice performance. Code will be made available at: www.github.com/mwyburd/TEDS-Net
Sli2Vol: Annotate a 3D Volume from a Single Slice with Self-Supervised Learning
May 26, 2021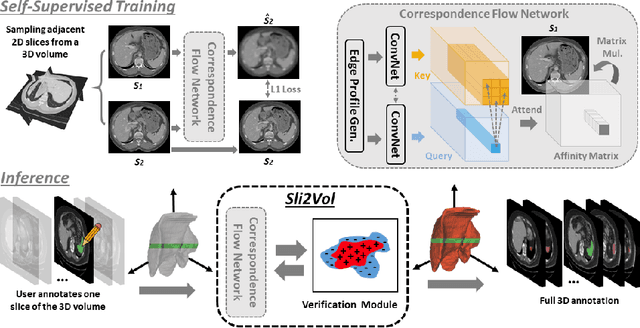
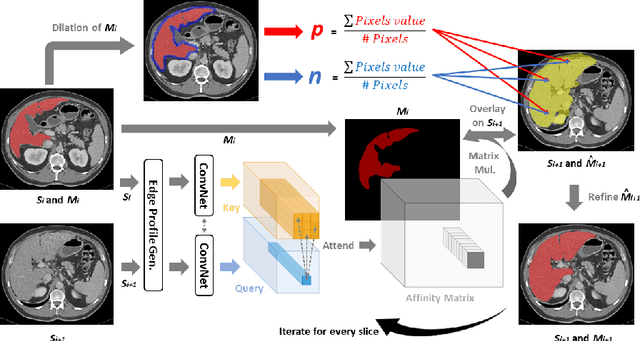
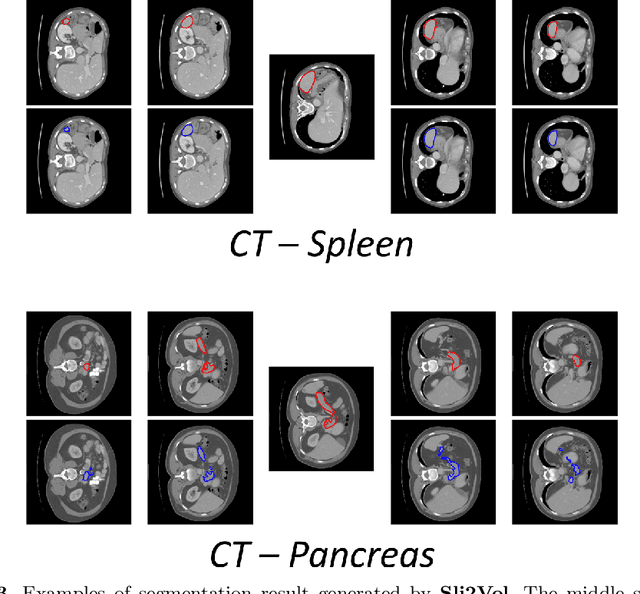
Abstract:The objective of this work is to segment any arbitrary structures of interest (SOI) in 3D volumes by only annotating a single slice, (i.e. semi-automatic 3D segmentation). We show that high accuracy can be achieved by simply propagating the 2D slice segmentation with an affinity matrix between consecutive slices, which can be learnt in a self-supervised manner, namely slice reconstruction. Specifically, we compare the proposed framework, termed as Sli2Vol, with supervised approaches and two other unsupervised/ self-supervised slice registration approaches, on 8 public datasets (both CT and MRI scans), spanning 9 different SOIs. Without any parameter-tuning, the same model achieves superior performance with Dice scores (0-100 scale) of over 80 for most of the benchmarks, including the ones that are unseen during training. Our results show generalizability of the proposed approach across data from different machines and with different SOIs: a major use case of semi-automatic segmentation methods where fully supervised approaches would normally struggle. The source code will be made publicly available at https://github.com/pakheiyeung/Sli2Vol.
Self-Supervised Ultrasound to MRI Fetal Brain Image Synthesis
Aug 19, 2020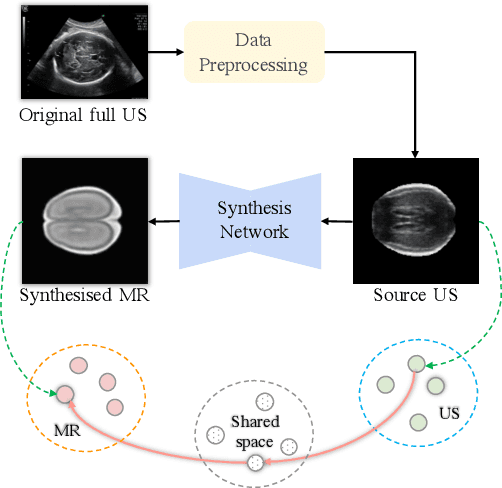
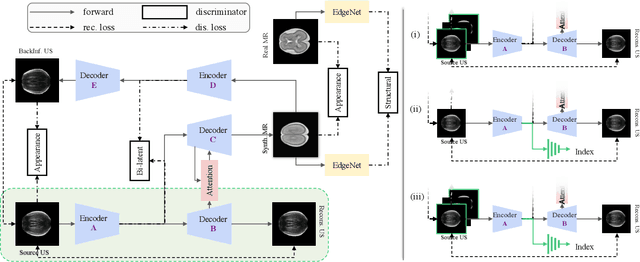
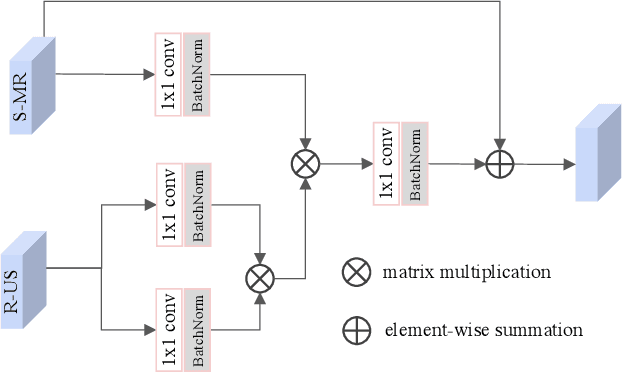
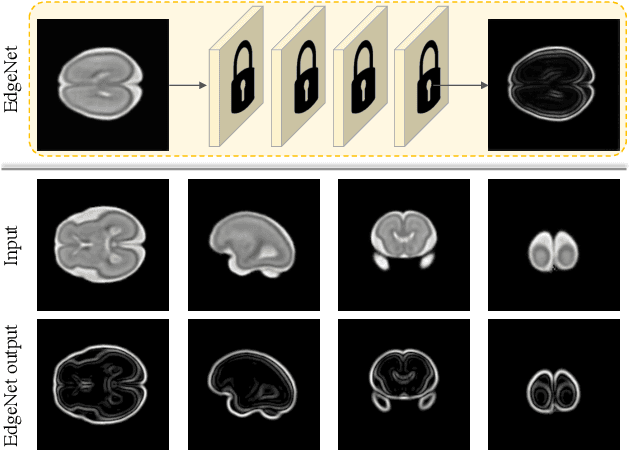
Abstract:Fetal brain magnetic resonance imaging (MRI) offers exquisite images of the developing brain but is not suitable for second-trimester anomaly screening, for which ultrasound (US) is employed. Although expert sonographers are adept at reading US images, MR images which closely resemble anatomical images are much easier for non-experts to interpret. Thus in this paper we propose to generate MR-like images directly from clinical US images. In medical image analysis such a capability is potentially useful as well, for instance for automatic US-MRI registration and fusion. The proposed model is end-to-end trainable and self-supervised without any external annotations. Specifically, based on an assumption that the US and MRI data share a similar anatomical latent space, we first utilise a network to extract the shared latent features, which are then used for MRI synthesis. Since paired data is unavailable for our study (and rare in practice), pixel-level constraints are infeasible to apply. We instead propose to enforce the distributions to be statistically indistinguishable, by adversarial learning in both the image domain and feature space. To regularise the anatomical structures between US and MRI during synthesis, we further propose an adversarial structural constraint. A new cross-modal attention technique is proposed to utilise non-local spatial information, by encouraging multi-modal knowledge fusion and propagation. We extend the approach to consider the case where 3D auxiliary information (e.g., 3D neighbours and a 3D location index) from volumetric data is also available, and show that this improves image synthesis. The proposed approach is evaluated quantitatively and qualitatively with comparison to real fetal MR images and other approaches to synthesis, demonstrating its feasibility of synthesising realistic MR images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge