Weidi Xie
Weaver: End-to-End Agentic System Training for Video Interleaved Reasoning
Feb 05, 2026Abstract:Video reasoning constitutes a comprehensive assessment of a model's capabilities, as it demands robust perceptual and interpretive skills, thereby serving as a means to explore the boundaries of model performance. While recent research has leveraged text-centric Chain-of-Thought reasoning to augment these capabilities, such approaches frequently suffer from representational mismatch and restricted by limited perceptual acuity. To address these limitations, we propose Weaver, a novel, end-to-end trainable multimodal reasoning agentic system. Weaver empowers its policy model to dynamically invoke diverse tools throughout the reasoning process, enabling progressive acquisition of crucial visual cues and construction of authentic multimodal reasoning trajectories. Furthermore, we integrate a reinforcement learning algorithm to allow the system to freely explore strategies for employing and combining these tools with trajectory-free data. Extensive experiments demonstrate that our system, Weaver, enhances performance on several complex video reasoning benchmarks, particularly those involving long videos.
Revisiting Multi-Task Visual Representation Learning
Jan 20, 2026Abstract:Current visual representation learning remains bifurcated: vision-language models (e.g., CLIP) excel at global semantic alignment but lack spatial precision, while self-supervised methods (e.g., MAE, DINO) capture intricate local structures yet struggle with high-level semantic context. We argue that these paradigms are fundamentally complementary and can be integrated into a principled multi-task framework, further enhanced by dense spatial supervision. We introduce MTV, a multi-task visual pretraining framework that jointly optimizes a shared backbone across vision-language contrastive, self-supervised, and dense spatial objectives. To mitigate the need for manual annotations, we leverage high-capacity "expert" models -- such as Depth Anything V2 and OWLv2 -- to synthesize dense, structured pseudo-labels at scale. Beyond the framework, we provide a systematic investigation into the mechanics of multi-task visual learning, analyzing: (i) the marginal gain of each objective, (ii) task synergies versus interference, and (iii) scaling behavior across varying data and model scales. Our results demonstrate that MTV achieves "best-of-both-worlds" performance, significantly enhancing fine-grained spatial reasoning without compromising global semantic understanding. Our findings suggest that multi-task learning, fueled by high-quality pseudo-supervision, is a scalable path toward more general visual encoders.
SoccerMaster: A Vision Foundation Model for Soccer Understanding
Dec 11, 2025Abstract:Soccer understanding has recently garnered growing research interest due to its domain-specific complexity and unique challenges. Unlike prior works that typically rely on isolated, task-specific expert models, this work aims to propose a unified model to handle diverse soccer visual understanding tasks, ranging from fine-grained perception (e.g., athlete detection) to semantic reasoning (e.g., event classification). Specifically, our contributions are threefold: (i) we present SoccerMaster, the first soccer-specific vision foundation model that unifies diverse understanding tasks within a single framework via supervised multi-task pretraining; (ii) we develop an automated data curation pipeline to generate scalable spatial annotations, and integrate them with various existing soccer video datasets to construct SoccerFactory, a comprehensive pretraining data resource; and (iii) we conduct extensive evaluations demonstrating that SoccerMaster consistently outperforms task-specific expert models across diverse downstream tasks, highlighting its breadth and superiority. The data, code, and model will be publicly available.
Inferring Dynamic Physical Properties from Video Foundation Models
Oct 02, 2025Abstract:We study the task of predicting dynamic physical properties from videos. More specifically, we consider physical properties that require temporal information to be inferred: elasticity of a bouncing object, viscosity of a flowing liquid, and dynamic friction of an object sliding on a surface. To this end, we make the following contributions: (i) We collect a new video dataset for each physical property, consisting of synthetic training and testing splits, as well as a real split for real world evaluation. (ii) We explore three ways to infer the physical property from videos: (a) an oracle method where we supply the visual cues that intrinsically reflect the property using classical computer vision techniques; (b) a simple read out mechanism using a visual prompt and trainable prompt vector for cross-attention on pre-trained video generative and self-supervised models; and (c) prompt strategies for Multi-modal Large Language Models (MLLMs). (iii) We show that video foundation models trained in a generative or self-supervised manner achieve a similar performance, though behind that of the oracle, and MLLMs are currently inferior to the other models, though their performance can be improved through suitable prompting.
Universal Video Temporal Grounding with Generative Multi-modal Large Language Models
Jun 23, 2025Abstract:This paper presents a computational model for universal video temporal grounding, which accurately localizes temporal moments in videos based on natural language queries (e.g., questions or descriptions). Unlike existing methods that are often limited to specific video domains or durations, we propose UniTime, a robust and universal video grounding model leveraging the strong vision-language understanding capabilities of generative Multi-modal Large Language Models (MLLMs). Our model effectively handles videos of diverse views, genres, and lengths while comprehending complex language queries. The key contributions include: (i) We consider steering strong MLLMs for temporal grounding in videos. To enable precise timestamp outputs, we incorporate temporal information by interleaving timestamp tokens with video tokens. (ii) By training the model to handle videos with different input granularities through adaptive frame scaling, our approach achieves robust temporal grounding for both short and long videos. (iii) Comprehensive experiments show that UniTime outperforms state-of-the-art approaches in both zero-shot and dataset-specific finetuned settings across five public temporal grounding benchmarks. (iv) When employed as a preliminary moment retriever for long-form video question-answering (VideoQA), UniTime significantly improves VideoQA accuracy, highlighting its value for complex video understanding tasks.
SpatialScore: Towards Unified Evaluation for Multimodal Spatial Understanding
May 22, 2025Abstract:Multimodal large language models (MLLMs) have achieved impressive success in question-answering tasks, yet their capabilities for spatial understanding are less explored. This work investigates a critical question: do existing MLLMs possess 3D spatial perception and understanding abilities? Concretely, we make the following contributions in this paper: (i) we introduce VGBench, a benchmark specifically designed to assess MLLMs for visual geometry perception, e.g., camera pose and motion estimation; (ii) we propose SpatialScore, the most comprehensive and diverse multimodal spatial understanding benchmark to date, integrating VGBench with relevant data from the other 11 existing datasets. This benchmark comprises 28K samples across various spatial understanding tasks, modalities, and QA formats, along with a carefully curated challenging subset, SpatialScore-Hard; (iii) we develop SpatialAgent, a novel multi-agent system incorporating 9 specialized tools for spatial understanding, supporting both Plan-Execute and ReAct reasoning paradigms; (iv) we conduct extensive evaluations to reveal persistent challenges in spatial reasoning while demonstrating the effectiveness of SpatialAgent. We believe SpatialScore will offer valuable insights and serve as a rigorous benchmark for the next evolution of MLLMs.
Multi-Agent System for Comprehensive Soccer Understanding
May 06, 2025Abstract:Recent advancements in AI-driven soccer understanding have demonstrated rapid progress, yet existing research predominantly focuses on isolated or narrow tasks. To bridge this gap, we propose a comprehensive framework for holistic soccer understanding. Specifically, we make the following contributions in this paper: (i) we construct SoccerWiki, the first large-scale multimodal soccer knowledge base, integrating rich domain knowledge about players, teams, referees, and venues to enable knowledge-driven reasoning; (ii) we present SoccerBench, the largest and most comprehensive soccer-specific benchmark, featuring around 10K standardized multimodal (text, image, video) multi-choice QA pairs across 13 distinct understanding tasks, curated through automated pipelines and manual verification; (iii) we introduce SoccerAgent, a novel multi-agent system that decomposes complex soccer questions via collaborative reasoning, leveraging domain expertise from SoccerWiki and achieving robust performance; (iv) extensive evaluations and ablations that benchmark state-of-the-art MLLMs on SoccerBench, highlighting the superiority of our proposed agentic system. All data and code are publicly available at: https://jyrao.github.io/SoccerAgent/.
ChestX-Reasoner: Advancing Radiology Foundation Models with Reasoning through Step-by-Step Verification
Apr 29, 2025Abstract:Recent advances in reasoning-enhanced large language models (LLMs) and multimodal LLMs (MLLMs) have significantly improved performance in complex tasks, yet medical AI models often overlook the structured reasoning processes inherent in clinical practice. In this work, we present ChestX-Reasoner, a radiology diagnosis MLLM designed to leverage process supervision mined directly from clinical reports, reflecting the step-by-step reasoning followed by radiologists. We construct a large dataset by extracting and refining reasoning chains from routine radiology reports. Our two-stage training framework combines supervised fine-tuning and reinforcement learning guided by process rewards to better align model reasoning with clinical standards. We introduce RadRBench-CXR, a comprehensive benchmark featuring 59K visual question answering samples with 301K clinically validated reasoning steps, and propose RadRScore, a metric evaluating reasoning factuality, completeness, and effectiveness. ChestX-Reasoner outperforms existing medical and general-domain MLLMs in both diagnostic accuracy and reasoning ability, achieving 16%, 5.9%, and 18% improvements in reasoning ability compared to the best medical MLLM, the best general MLLM, and its base model, respectively, as well as 3.3%, 24%, and 27% improvements in outcome accuracy. All resources are open-sourced to facilitate further research in medical reasoning MLLMs.
Learning Streaming Video Representation via Multitask Training
Apr 28, 2025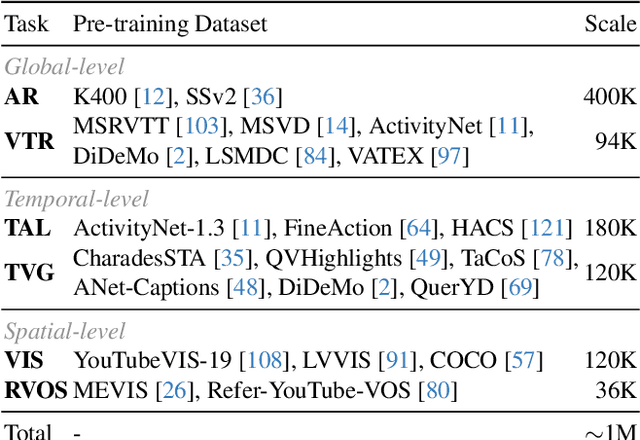
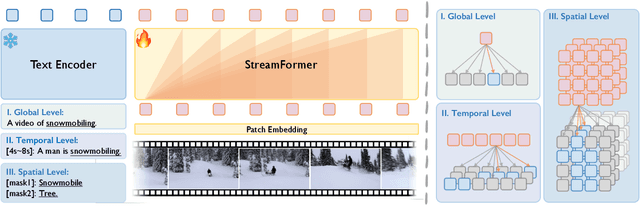
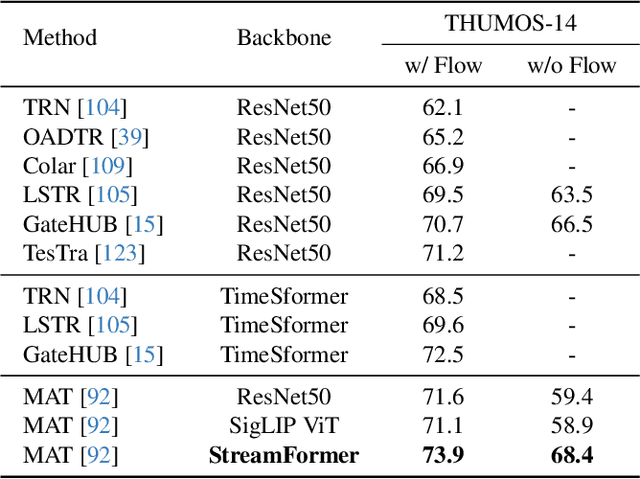
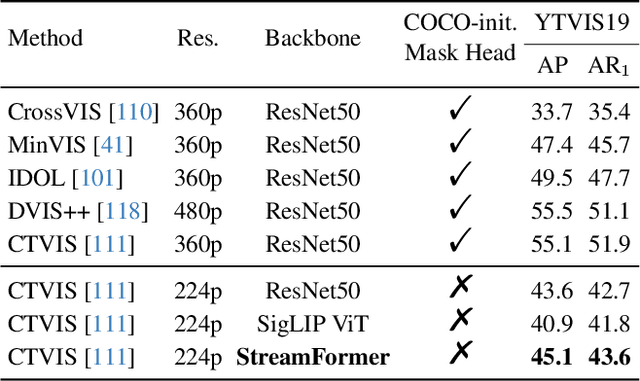
Abstract:Understanding continuous video streams plays a fundamental role in real-time applications including embodied AI and autonomous driving. Unlike offline video understanding, streaming video understanding requires the ability to process video streams frame by frame, preserve historical information, and make low-latency decisions.To address these challenges, our main contributions are three-fold. (i) We develop a novel streaming video backbone, termed as StreamFormer, by incorporating causal temporal attention into a pre-trained vision transformer. This enables efficient streaming video processing while maintaining image representation capability.(ii) To train StreamFormer, we propose to unify diverse spatial-temporal video understanding tasks within a multitask visual-language alignment framework. Hence, StreamFormer learns global semantics, temporal dynamics, and fine-grained spatial relationships simultaneously. (iii) We conduct extensive experiments on online action detection, online video instance segmentation, and video question answering. StreamFormer achieves competitive results while maintaining efficiency, demonstrating its potential for real-time applications.
EgoExo-Gen: Ego-centric Video Prediction by Watching Exo-centric Videos
Apr 16, 2025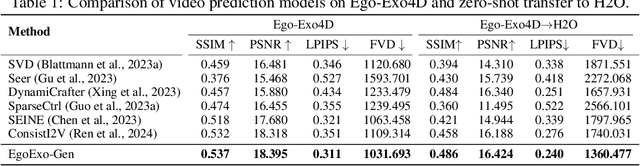
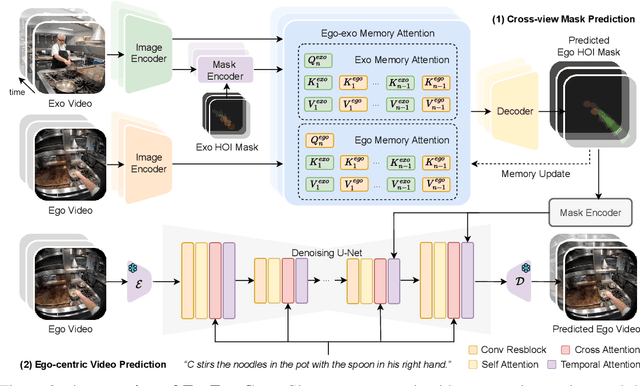
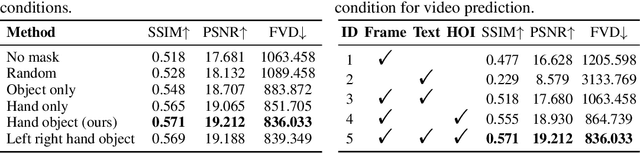
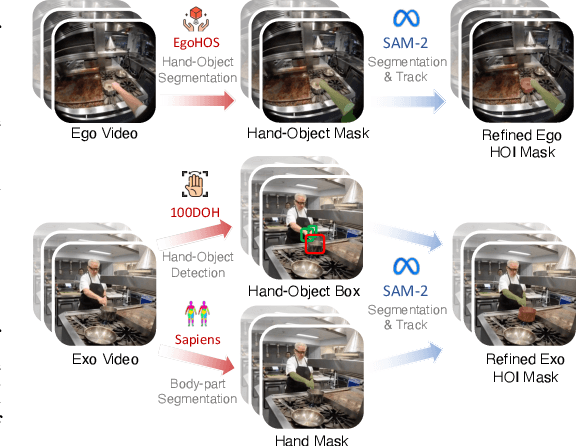
Abstract:Generating videos in the first-person perspective has broad application prospects in the field of augmented reality and embodied intelligence. In this work, we explore the cross-view video prediction task, where given an exo-centric video, the first frame of the corresponding ego-centric video, and textual instructions, the goal is to generate futur frames of the ego-centric video. Inspired by the notion that hand-object interactions (HOI) in ego-centric videos represent the primary intentions and actions of the current actor, we present EgoExo-Gen that explicitly models the hand-object dynamics for cross-view video prediction. EgoExo-Gen consists of two stages. First, we design a cross-view HOI mask prediction model that anticipates the HOI masks in future ego-frames by modeling the spatio-temporal ego-exo correspondence. Next, we employ a video diffusion model to predict future ego-frames using the first ego-frame and textual instructions, while incorporating the HOI masks as structural guidance to enhance prediction quality. To facilitate training, we develop an automated pipeline to generate pseudo HOI masks for both ego- and exo-videos by exploiting vision foundation models. Extensive experiments demonstrate that our proposed EgoExo-Gen achieves better prediction performance compared to previous video prediction models on the Ego-Exo4D and H2O benchmark datasets, with the HOI masks significantly improving the generation of hands and interactive objects in the ego-centric videos.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge