Chaoyi Wu
ChestX-Reasoner: Advancing Radiology Foundation Models with Reasoning through Step-by-Step Verification
Apr 29, 2025Abstract:Recent advances in reasoning-enhanced large language models (LLMs) and multimodal LLMs (MLLMs) have significantly improved performance in complex tasks, yet medical AI models often overlook the structured reasoning processes inherent in clinical practice. In this work, we present ChestX-Reasoner, a radiology diagnosis MLLM designed to leverage process supervision mined directly from clinical reports, reflecting the step-by-step reasoning followed by radiologists. We construct a large dataset by extracting and refining reasoning chains from routine radiology reports. Our two-stage training framework combines supervised fine-tuning and reinforcement learning guided by process rewards to better align model reasoning with clinical standards. We introduce RadRBench-CXR, a comprehensive benchmark featuring 59K visual question answering samples with 301K clinically validated reasoning steps, and propose RadRScore, a metric evaluating reasoning factuality, completeness, and effectiveness. ChestX-Reasoner outperforms existing medical and general-domain MLLMs in both diagnostic accuracy and reasoning ability, achieving 16%, 5.9%, and 18% improvements in reasoning ability compared to the best medical MLLM, the best general MLLM, and its base model, respectively, as well as 3.3%, 24%, and 27% improvements in outcome accuracy. All resources are open-sourced to facilitate further research in medical reasoning MLLMs.
Quantifying the Reasoning Abilities of LLMs on Real-world Clinical Cases
Mar 06, 2025

Abstract:The latest reasoning-enhanced large language models (reasoning LLMs), such as DeepSeek-R1 and OpenAI-o3, have demonstrated remarkable success. However, the application of such reasoning enhancements to the highly professional medical domain has not been clearly evaluated, particularly regarding with not only assessing the final generation but also examining the quality of their reasoning processes. In this study, we present MedR-Bench, a reasoning-focused medical evaluation benchmark comprising 1,453 structured patient cases with reasoning references mined from case reports. Our benchmark spans 13 body systems and 10 specialty disorders, encompassing both common and rare diseases. In our evaluation, we introduce a versatile framework consisting of three critical clinical stages: assessment recommendation, diagnostic decision-making, and treatment planning, comprehensively capturing the LLMs' performance across the entire patient journey in healthcare. For metrics, we propose a novel agentic system, Reasoning Evaluator, designed to automate and objectively quantify free-text reasoning responses in a scalable manner from the perspectives of efficiency, factuality, and completeness by dynamically searching and performing cross-referencing checks. As a result, we assess five state-of-the-art reasoning LLMs, including DeepSeek-R1, OpenAI-o3-mini, and others. Our results reveal that current LLMs can handle relatively simple diagnostic tasks with sufficient critical assessment results, achieving accuracy generally over 85%. However, they still struggle with more complex tasks, such as assessment recommendation and treatment planning. In reasoning, their reasoning processes are generally reliable, with factuality scores exceeding 90%, though they often omit critical reasoning steps. Our study clearly reveals further development directions for current clinical LLMs.
RadIR: A Scalable Framework for Multi-Grained Medical Image Retrieval via Radiology Report Mining
Mar 06, 2025Abstract:Developing advanced medical imaging retrieval systems is challenging due to the varying definitions of `similar images' across different medical contexts. This challenge is compounded by the lack of large-scale, high-quality medical imaging retrieval datasets and benchmarks. In this paper, we propose a novel methodology that leverages dense radiology reports to define image-wise similarity ordering at multiple granularities in a scalable and fully automatic manner. Using this approach, we construct two comprehensive medical imaging retrieval datasets: MIMIC-IR for Chest X-rays and CTRATE-IR for CT scans, providing detailed image-image ranking annotations conditioned on diverse anatomical structures. Furthermore, we develop two retrieval systems, RadIR-CXR and model-ChestCT, which demonstrate superior performance in traditional image-image and image-report retrieval tasks. These systems also enable flexible, effective image retrieval conditioned on specific anatomical structures described in text, achieving state-of-the-art results on 77 out of 78 metrics.
M^3Builder: A Multi-Agent System for Automated Machine Learning in Medical Imaging
Feb 27, 2025Abstract:Agentic AI systems have gained significant attention for their ability to autonomously perform complex tasks. However, their reliance on well-prepared tools limits their applicability in the medical domain, which requires to train specialized models. In this paper, we make three contributions: (i) We present M3Builder, a novel multi-agent system designed to automate machine learning (ML) in medical imaging. At its core, M3Builder employs four specialized agents that collaborate to tackle complex, multi-step medical ML workflows, from automated data processing and environment configuration to self-contained auto debugging and model training. These agents operate within a medical imaging ML workspace, a structured environment designed to provide agents with free-text descriptions of datasets, training codes, and interaction tools, enabling seamless communication and task execution. (ii) To evaluate progress in automated medical imaging ML, we propose M3Bench, a benchmark comprising four general tasks on 14 training datasets, across five anatomies and three imaging modalities, covering both 2D and 3D data. (iii) We experiment with seven state-of-the-art large language models serving as agent cores for our system, such as Claude series, GPT-4o, and DeepSeek-V3. Compared to existing ML agentic designs, M3Builder shows superior performance on completing ML tasks in medical imaging, achieving a 94.29% success rate using Claude-3.7-Sonnet as the agent core, showing huge potential towards fully automated machine learning in medical imaging.
Can Modern LLMs Act as Agent Cores in Radiology~Environments?
Dec 12, 2024Abstract:Advancements in large language models (LLMs) have paved the way for LLM-based agent systems that offer enhanced accuracy and interpretability across various domains. Radiology, with its complex analytical requirements, is an ideal field for the application of these agents. This paper aims to investigate the pre-requisite question for building concrete radiology agents which is, `Can modern LLMs act as agent cores in radiology environments?' To investigate it, we introduce RadABench with three-fold contributions: First, we present RadABench-Data, a comprehensive synthetic evaluation dataset for LLM-based agents, generated from an extensive taxonomy encompassing 6 anatomies, 5 imaging modalities, 10 tool categories, and 11 radiology tasks. Second, we propose RadABench-EvalPlat, a novel evaluation platform for agents featuring a prompt-driven workflow and the capability to simulate a wide range of radiology toolsets. Third, we assess the performance of 7 leading LLMs on our benchmark from 5 perspectives with multiple metrics. Our findings indicate that while current LLMs demonstrate strong capabilities in many areas, they are still not sufficiently advanced to serve as the central agent core in a fully operational radiology agent system. Additionally, we identify key factors influencing the performance of LLM-based agent cores, offering insights for clinicians on how to apply agent systems in real-world radiology practices effectively. All of our code and data are open-sourced in https://github.com/MAGIC-AI4Med/RadABench.
Towards Evaluating and Building Versatile Large Language Models for Medicine
Aug 22, 2024Abstract:In this study, we present MedS-Bench, a comprehensive benchmark designed to evaluate the performance of large language models (LLMs) in clinical contexts. Unlike existing benchmarks that focus on multiple-choice question answering, MedS-Bench spans 11 high-level clinical tasks, including clinical report summarization, treatment recommendations, diagnosis, named entity recognition, and medical concept explanation, among others. We evaluated six leading LLMs, e.g., MEDITRON, Mistral, InternLM 2, Llama 3, GPT-4, and Claude-3.5 using few-shot prompting, and found that even the most sophisticated models struggle with these complex tasks. To address these limitations, we developed MedS-Ins, a large-scale instruction tuning dataset for medicine. MedS-Ins comprises 58 medically oriented language corpora, totaling 13.5 million samples across 122 tasks. To demonstrate the dataset's utility, we conducted a proof-of-concept experiment by performing instruction tuning on a lightweight, open-source medical language model. The resulting model, MMedIns-Llama 3, significantly outperformed existing models across nearly all clinical tasks. To promote further advancements in the application of LLMs to clinical challenges, we have made the MedS-Ins dataset fully accessible and invite the research community to contribute to its expansion.Additionally, we have launched a dynamic leaderboard for MedS-Bench, which we plan to regularly update the test set to track progress and enhance the adaptation of general LLMs to the medical domain. Leaderboard: https://henrychur.github.io/MedS-Bench/. Github: https://github.com/MAGIC-AI4Med/MedS-Ins.
AutoRG-Brain: Grounded Report Generation for Brain MRI
Jul 26, 2024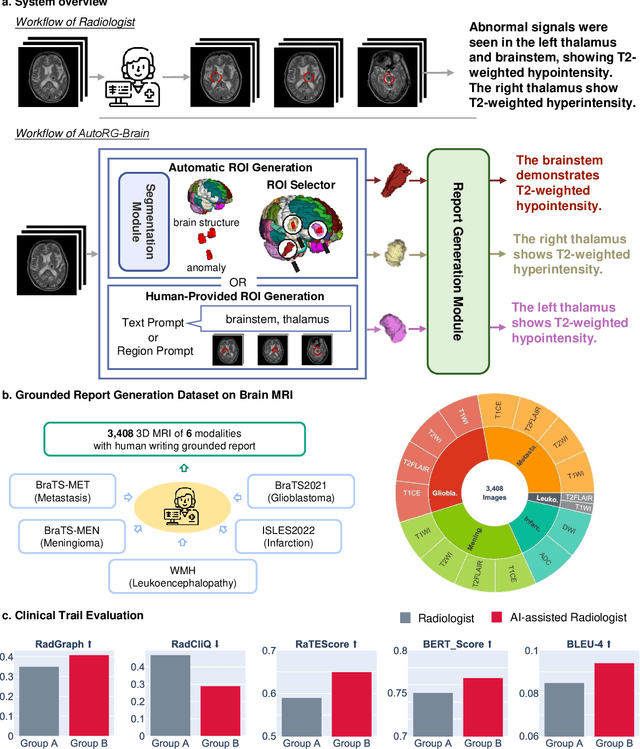
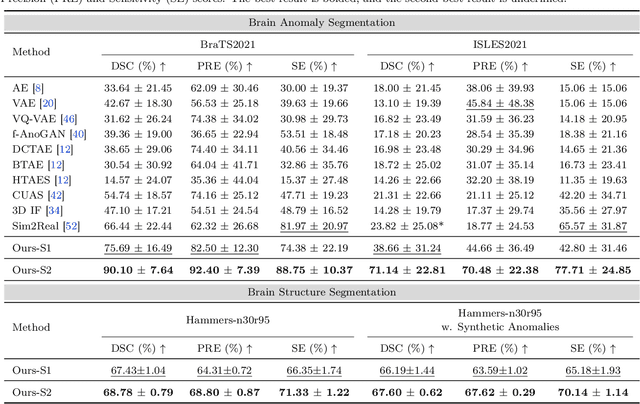
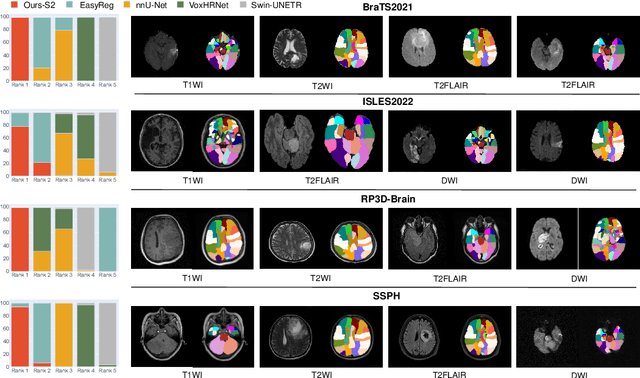
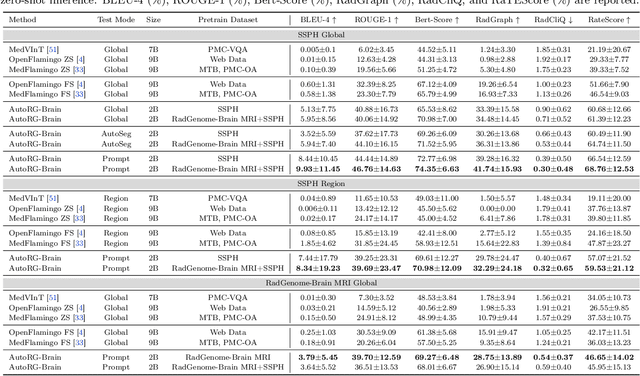
Abstract:Radiologists are tasked with interpreting a large number of images in a daily base, with the responsibility of generating corresponding reports. This demanding workload elevates the risk of human error, potentially leading to treatment delays, increased healthcare costs, revenue loss, and operational inefficiencies. To address these challenges, we initiate a series of work on grounded Automatic Report Generation (AutoRG), starting from the brain MRI interpretation system, which supports the delineation of brain structures, the localization of anomalies, and the generation of well-organized findings. We make contributions from the following aspects, first, on dataset construction, we release a comprehensive dataset encompassing segmentation masks of anomaly regions and manually authored reports, termed as RadGenome-Brain MRI. This data resource is intended to catalyze ongoing research and development in the field of AI-assisted report generation systems. Second, on system design, we propose AutoRG-Brain, the first brain MRI report generation system with pixel-level grounded visual clues. Third, for evaluation, we conduct quantitative assessments and human evaluations of brain structure segmentation, anomaly localization, and report generation tasks to provide evidence of its reliability and accuracy. This system has been integrated into real clinical scenarios, where radiologists were instructed to write reports based on our generated findings and anomaly segmentation masks. The results demonstrate that our system enhances the report-writing skills of junior doctors, aligning their performance more closely with senior doctors, thereby boosting overall productivity.
RaTEScore: A Metric for Radiology Report Generation
Jun 24, 2024Abstract:This paper introduces a novel, entity-aware metric, termed as Radiological Report (Text) Evaluation (RaTEScore), to assess the quality of medical reports generated by AI models. RaTEScore emphasizes crucial medical entities such as diagnostic outcomes and anatomical details, and is robust against complex medical synonyms and sensitive to negation expressions. Technically, we developed a comprehensive medical NER dataset, RaTE-NER, and trained an NER model specifically for this purpose. This model enables the decomposition of complex radiological reports into constituent medical entities. The metric itself is derived by comparing the similarity of entity embeddings, obtained from a language model, based on their types and relevance to clinical significance. Our evaluations demonstrate that RaTEScore aligns more closely with human preference than existing metrics, validated both on established public benchmarks and our newly proposed RaTE-Eval benchmark.
RadGenome-Chest CT: A Grounded Vision-Language Dataset for Chest CT Analysis
Apr 25, 2024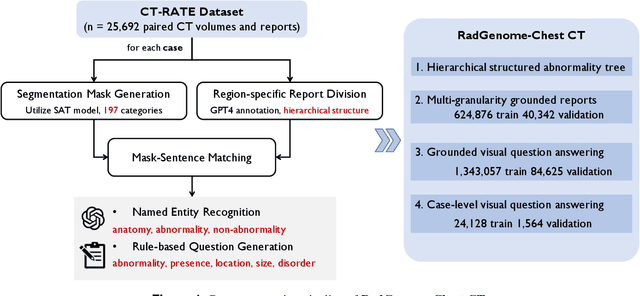


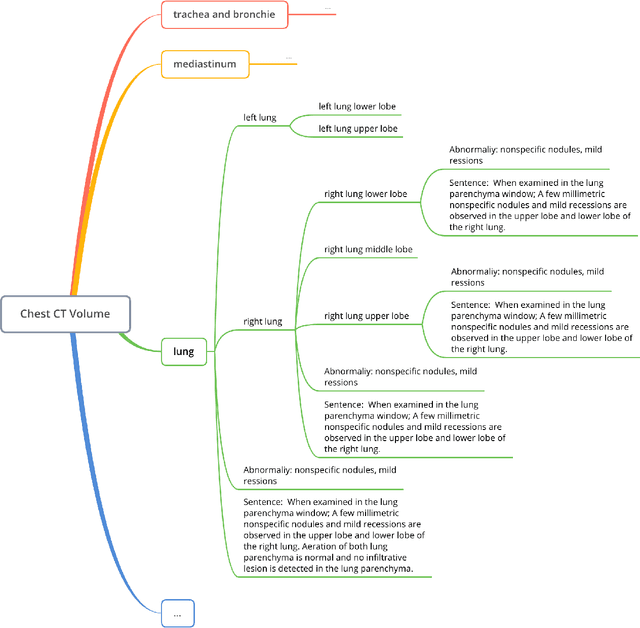
Abstract:Developing generalist foundation model has recently attracted tremendous attention among researchers in the field of AI for Medicine (AI4Medicine). A pivotal insight in developing these models is their reliance on dataset scaling, which emphasizes the requirements on developing open-source medical image datasets that incorporate diverse supervision signals across various imaging modalities. In this paper, we introduce RadGenome-Chest CT, a comprehensive, large-scale, region-guided 3D chest CT interpretation dataset based on CT-RATE. Specifically, we leverage the latest powerful universal segmentation and large language models, to extend the original datasets (over 25,692 non-contrast 3D chest CT volume and reports from 20,000 patients) from the following aspects: (i) organ-level segmentation masks covering 197 categories, which provide intermediate reasoning visual clues for interpretation; (ii) 665 K multi-granularity grounded reports, where each sentence of the report is linked to the corresponding anatomical region of CT volume in the form of a segmentation mask; (iii) 1.3 M grounded VQA pairs, where questions and answers are all linked with reference segmentation masks, enabling models to associate visual evidence with textual explanations. All grounded reports and VQA pairs in the validation set have gone through manual verification to ensure dataset quality. We believe that RadGenome-Chest CT can significantly advance the development of multimodal medical foundation models, by training to generate texts based on given segmentation regions, which is unattainable with previous relevant datasets. We will release all segmentation masks, grounded reports, and VQA pairs to facilitate further research and development in this field.
Knowledge-enhanced Visual-Language Pretraining for Computational Pathology
Apr 15, 2024



Abstract:In this paper, we consider the problem of visual representation learning for computational pathology, by exploiting large-scale image-text pairs gathered from public resources, along with the domain specific knowledge in pathology. Specifically, we make the following contributions: (i) We curate a pathology knowledge tree that consists of 50,470 informative attributes for 4,718 diseases requiring pathology diagnosis from 32 human tissues. To our knowledge, this is the first comprehensive structured pathology knowledge base; (ii) We develop a knowledge-enhanced visual-language pretraining approach, where we first project pathology-specific knowledge into latent embedding space via language model, and use it to guide the visual representation learning; (iii) We conduct thorough experiments to validate the effectiveness of our proposed components, demonstrating significant performance improvement on various downstream tasks, including cross-modal retrieval, zero-shot classification on pathology patches, and zero-shot tumor subtyping on whole slide images (WSIs). All codes, models and the pathology knowledge tree will be released to the research community
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge