Yuehua Li
Open-World 3D Scene Graph Generation for Retrieval-Augmented Reasoning
Nov 08, 2025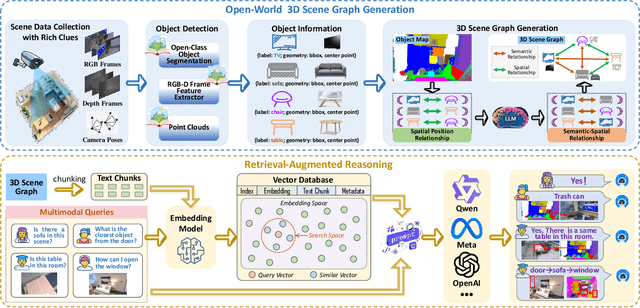
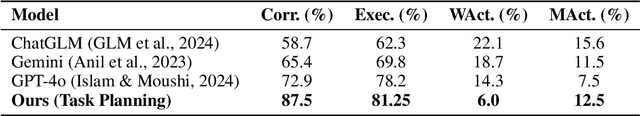
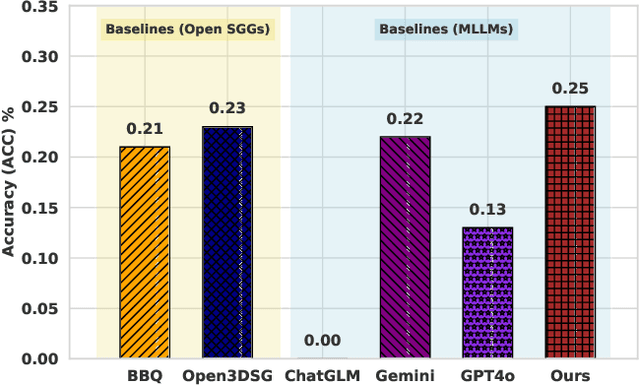
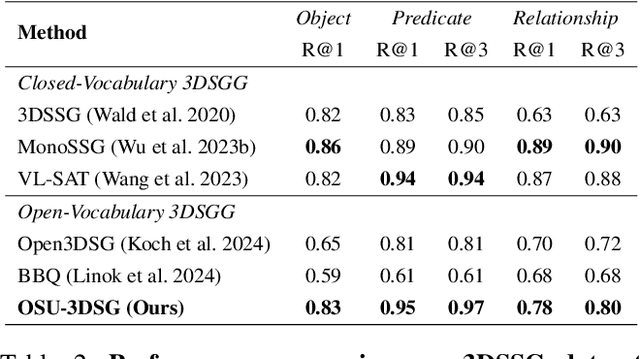
Abstract:Understanding 3D scenes in open-world settings poses fundamental challenges for vision and robotics, particularly due to the limitations of closed-vocabulary supervision and static annotations. To address this, we propose a unified framework for Open-World 3D Scene Graph Generation with Retrieval-Augmented Reasoning, which enables generalizable and interactive 3D scene understanding. Our method integrates Vision-Language Models (VLMs) with retrieval-based reasoning to support multimodal exploration and language-guided interaction. The framework comprises two key components: (1) a dynamic scene graph generation module that detects objects and infers semantic relationships without fixed label sets, and (2) a retrieval-augmented reasoning pipeline that encodes scene graphs into a vector database to support text/image-conditioned queries. We evaluate our method on 3DSSG and Replica benchmarks across four tasks-scene question answering, visual grounding, instance retrieval, and task planning-demonstrating robust generalization and superior performance in diverse environments. Our results highlight the effectiveness of combining open-vocabulary perception with retrieval-based reasoning for scalable 3D scene understanding.
Enhanced Multimodal RAG-LLM for Accurate Visual Question Answering
Dec 30, 2024



Abstract:Multimodal large language models (MLLMs), such as GPT-4o, Gemini, LLaVA, and Flamingo, have made significant progress in integrating visual and textual modalities, excelling in tasks like visual question answering (VQA), image captioning, and content retrieval. They can generate coherent and contextually relevant descriptions of images. However, they still face challenges in accurately identifying and counting objects and determining their spatial locations, particularly in complex scenes with overlapping or small objects. To address these limitations, we propose a novel framework based on multimodal retrieval-augmented generation (RAG), which introduces structured scene graphs to enhance object recognition, relationship identification, and spatial understanding within images. Our framework improves the MLLM's capacity to handle tasks requiring precise visual descriptions, especially in scenarios with challenging perspectives, such as aerial views or scenes with dense object arrangements. Finally, we conduct extensive experiments on the VG-150 dataset that focuses on first-person visual understanding and the AUG dataset that involves aerial imagery. The results show that our approach consistently outperforms existing MLLMs in VQA tasks, which stands out in recognizing, localizing, and quantifying objects in different spatial contexts and provides more accurate visual descriptions.
AutoRG-Brain: Grounded Report Generation for Brain MRI
Jul 26, 2024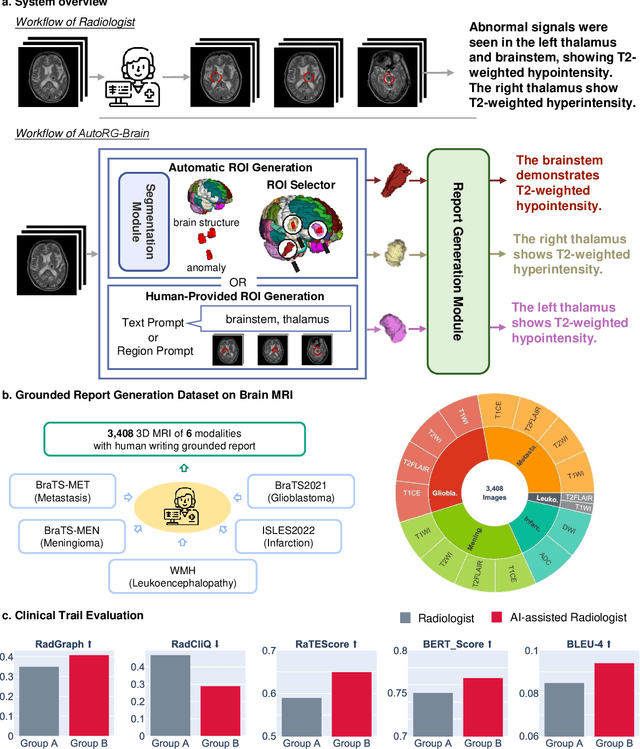
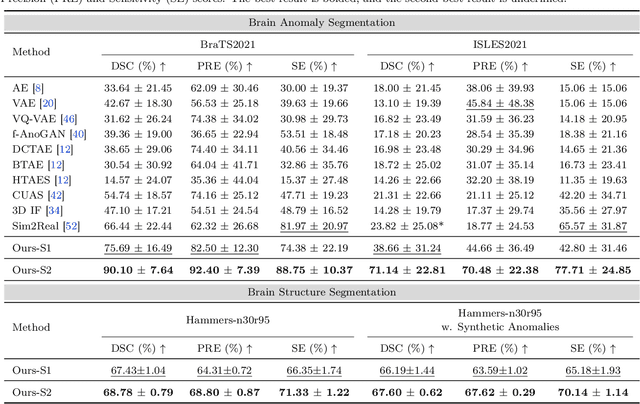
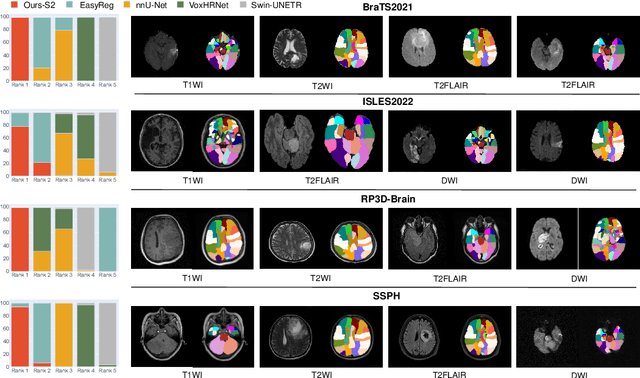
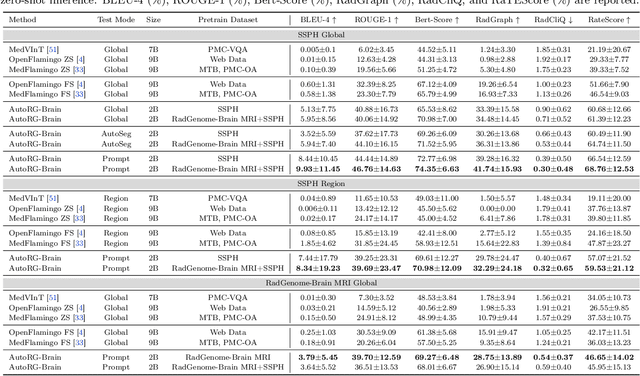
Abstract:Radiologists are tasked with interpreting a large number of images in a daily base, with the responsibility of generating corresponding reports. This demanding workload elevates the risk of human error, potentially leading to treatment delays, increased healthcare costs, revenue loss, and operational inefficiencies. To address these challenges, we initiate a series of work on grounded Automatic Report Generation (AutoRG), starting from the brain MRI interpretation system, which supports the delineation of brain structures, the localization of anomalies, and the generation of well-organized findings. We make contributions from the following aspects, first, on dataset construction, we release a comprehensive dataset encompassing segmentation masks of anomaly regions and manually authored reports, termed as RadGenome-Brain MRI. This data resource is intended to catalyze ongoing research and development in the field of AI-assisted report generation systems. Second, on system design, we propose AutoRG-Brain, the first brain MRI report generation system with pixel-level grounded visual clues. Third, for evaluation, we conduct quantitative assessments and human evaluations of brain structure segmentation, anomaly localization, and report generation tasks to provide evidence of its reliability and accuracy. This system has been integrated into real clinical scenarios, where radiologists were instructed to write reports based on our generated findings and anomaly segmentation masks. The results demonstrate that our system enhances the report-writing skills of junior doctors, aligning their performance more closely with senior doctors, thereby boosting overall productivity.
UniBrain: Universal Brain MRI Diagnosis with Hierarchical Knowledge-enhanced Pre-training
Sep 13, 2023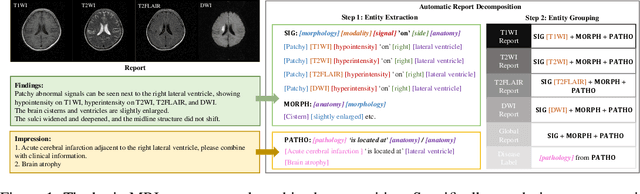
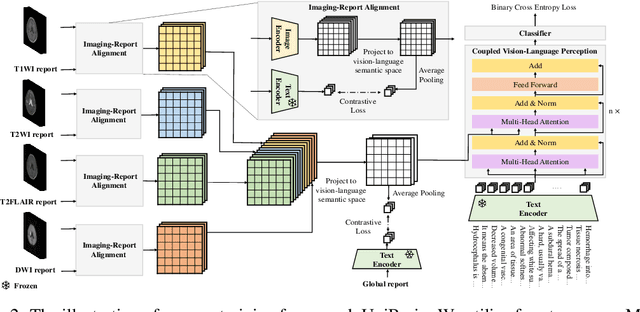
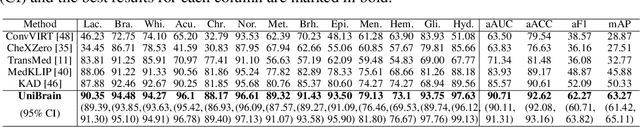
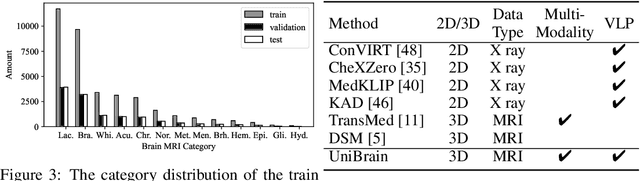
Abstract:Magnetic resonance imaging~(MRI) have played a crucial role in brain disease diagnosis, with which a range of computer-aided artificial intelligence methods have been proposed. However, the early explorations usually focus on the limited types of brain diseases in one study and train the model on the data in a small scale, yielding the bottleneck of generalization. Towards a more effective and scalable paradigm, we propose a hierarchical knowledge-enhanced pre-training framework for the universal brain MRI diagnosis, termed as UniBrain. Specifically, UniBrain leverages a large-scale dataset of 24,770 imaging-report pairs from routine diagnostics. Different from previous pre-training techniques for the unitary vision or textual feature, or with the brute-force alignment between vision and language information, we leverage the unique characteristic of report information in different granularity to build a hierarchical alignment mechanism, which strengthens the efficiency in feature learning. Our UniBrain is validated on three real world datasets with severe class imbalance and the public BraTS2019 dataset. It not only consistently outperforms all state-of-the-art diagnostic methods by a large margin and provides a superior grounding performance but also shows comparable performance compared to expert radiologists on certain disease types.
Two-Stream Compare and Contrast Network for Vertebral Compression Fracture Diagnosis
Oct 13, 2020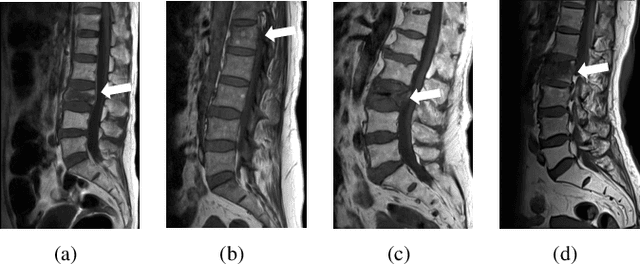
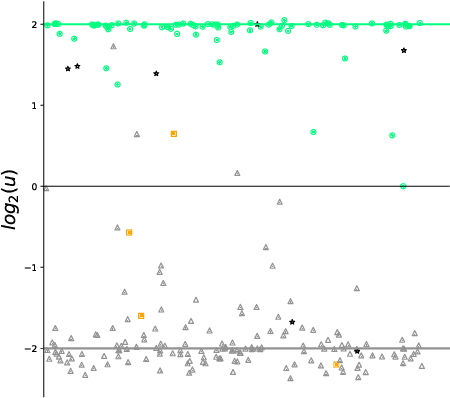
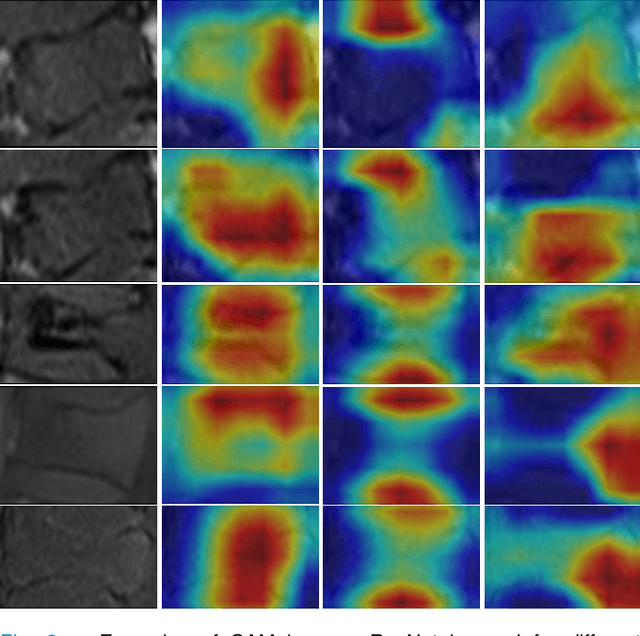
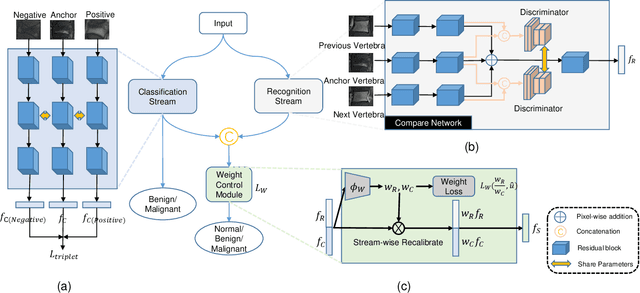
Abstract:Differentiating Vertebral Compression Fractures (VCFs) associated with trauma and osteoporosis (benign VCFs) or those caused by metastatic cancer (malignant VCFs) are critically important for treatment decisions. So far, automatic VCFs diagnosis is solved in a two-step manner, i.e. first identify VCFs and then classify it into benign or malignant. In this paper, we explore to model VCFs diagnosis as a three-class classification problem, i.e. normal vertebrae, benign VCFs, and malignant VCFs. However, VCFs recognition and classification require very different features, and both tasks are characterized by high intra-class variation and high inter-class similarity. Moreover, the dataset is extremely class-imbalanced. To address the above challenges, we propose a novel Two-Stream Compare and Contrast Network (TSCCN) for VCFs diagnosis. This network consists of two streams, a recognition stream which learns to identify VCFs through comparing and contrasting between adjacent vertebra, and a classification stream which compares and contrasts between intra-class and inter-class to learn features for fine-grained classification. The two streams are integrated via a learnable weight control module which adaptively sets their contribution. The TSCCN is evaluated on a dataset consisting of 239 VCFs patients and achieves the average sensitivity and specificity of 92.56\% and 96.29\%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge