Shixiang Feng
MS-KD: Multi-Organ Segmentation with Multiple Binary-Labeled Datasets
Aug 05, 2021



Abstract:Annotating multiple organs in 3D medical images is time-consuming and costly. Meanwhile, there exist many single-organ datasets with one specific organ annotated. This paper investigates how to learn a multi-organ segmentation model leveraging a set of binary-labeled datasets. A novel Multi-teacher Single-student Knowledge Distillation (MS-KD) framework is proposed, where the teacher models are pre-trained single-organ segmentation networks, and the student model is a multi-organ segmentation network. Considering that each teacher focuses on different organs, a region-based supervision method, consisting of logits-wise supervision and feature-wise supervision, is proposed. Each teacher supervises the student in two regions, the organ region where the teacher is considered as an expert and the background region where all teachers agree. Extensive experiments on three public single-organ datasets and a multi-organ dataset have demonstrated the effectiveness of the proposed MS-KD framework.
Uncertainty-aware Incremental Learning for Multi-organ Segmentation
Mar 09, 2021



Abstract:Most existing approaches to train a unified multi-organ segmentation model from several single-organ datasets require simultaneously access multiple datasets during training. In the real scenarios, due to privacy and ethics concerns, the training data of the organs of interest may not be publicly available. To this end, we investigate a data-free incremental organ segmentation scenario and propose a novel incremental training framework to solve it. We use the pretrained model instead of its own training data for privacy protection. Specifically, given a pretrained $K$ organ segmentation model and a new single-organ dataset, we train a unified $K+1$ organ segmentation model without accessing any data belonging to the previous training stages. Our approach consists of two parts: the background label alignment strategy and the uncertainty-aware guidance strategy. The first part is used for knowledge transfer from the pretained model to the training model. The second part is used to extract the uncertainty information from the pretrained model to guide the whole knowledge transfer process. By combing these two strategies, more reliable information is extracted from the pretrained model without original training data. Experiments on multiple publicly available pretrained models and a multi-organ dataset MOBA have demonstrated the effectiveness of our framework.
Two-Stream Compare and Contrast Network for Vertebral Compression Fracture Diagnosis
Oct 13, 2020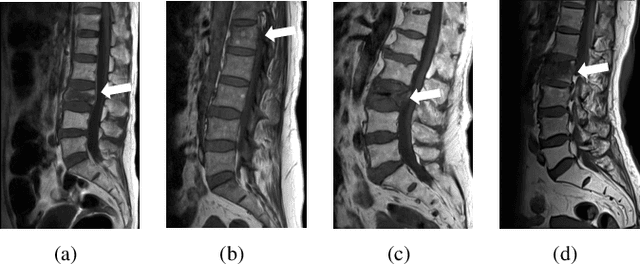
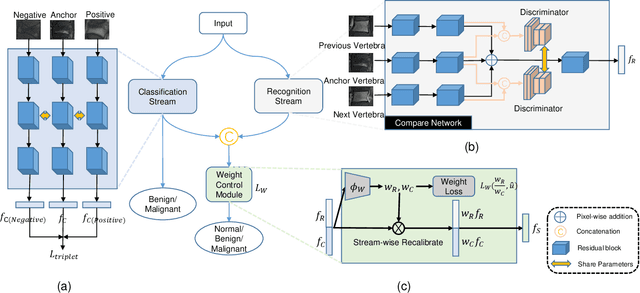
Abstract:Differentiating Vertebral Compression Fractures (VCFs) associated with trauma and osteoporosis (benign VCFs) or those caused by metastatic cancer (malignant VCFs) are critically important for treatment decisions. So far, automatic VCFs diagnosis is solved in a two-step manner, i.e. first identify VCFs and then classify it into benign or malignant. In this paper, we explore to model VCFs diagnosis as a three-class classification problem, i.e. normal vertebrae, benign VCFs, and malignant VCFs. However, VCFs recognition and classification require very different features, and both tasks are characterized by high intra-class variation and high inter-class similarity. Moreover, the dataset is extremely class-imbalanced. To address the above challenges, we propose a novel Two-Stream Compare and Contrast Network (TSCCN) for VCFs diagnosis. This network consists of two streams, a recognition stream which learns to identify VCFs through comparing and contrasting between adjacent vertebra, and a classification stream which compares and contrasts between intra-class and inter-class to learn features for fine-grained classification. The two streams are integrated via a learnable weight control module which adaptively sets their contribution. The TSCCN is evaluated on a dataset consisting of 239 VCFs patients and achieves the average sensitivity and specificity of 92.56\% and 96.29\%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge