Ziheng Zhao
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
RadIR: A Scalable Framework for Multi-Grained Medical Image Retrieval via Radiology Report Mining
Mar 06, 2025Abstract:Developing advanced medical imaging retrieval systems is challenging due to the varying definitions of `similar images' across different medical contexts. This challenge is compounded by the lack of large-scale, high-quality medical imaging retrieval datasets and benchmarks. In this paper, we propose a novel methodology that leverages dense radiology reports to define image-wise similarity ordering at multiple granularities in a scalable and fully automatic manner. Using this approach, we construct two comprehensive medical imaging retrieval datasets: MIMIC-IR for Chest X-rays and CTRATE-IR for CT scans, providing detailed image-image ranking annotations conditioned on diverse anatomical structures. Furthermore, we develop two retrieval systems, RadIR-CXR and model-ChestCT, which demonstrate superior performance in traditional image-image and image-report retrieval tasks. These systems also enable flexible, effective image retrieval conditioned on specific anatomical structures described in text, achieving state-of-the-art results on 77 out of 78 metrics.
M^3Builder: A Multi-Agent System for Automated Machine Learning in Medical Imaging
Feb 27, 2025Abstract:Agentic AI systems have gained significant attention for their ability to autonomously perform complex tasks. However, their reliance on well-prepared tools limits their applicability in the medical domain, which requires to train specialized models. In this paper, we make three contributions: (i) We present M3Builder, a novel multi-agent system designed to automate machine learning (ML) in medical imaging. At its core, M3Builder employs four specialized agents that collaborate to tackle complex, multi-step medical ML workflows, from automated data processing and environment configuration to self-contained auto debugging and model training. These agents operate within a medical imaging ML workspace, a structured environment designed to provide agents with free-text descriptions of datasets, training codes, and interaction tools, enabling seamless communication and task execution. (ii) To evaluate progress in automated medical imaging ML, we propose M3Bench, a benchmark comprising four general tasks on 14 training datasets, across five anatomies and three imaging modalities, covering both 2D and 3D data. (iii) We experiment with seven state-of-the-art large language models serving as agent cores for our system, such as Claude series, GPT-4o, and DeepSeek-V3. Compared to existing ML agentic designs, M3Builder shows superior performance on completing ML tasks in medical imaging, achieving a 94.29% success rate using Claude-3.7-Sonnet as the agent core, showing huge potential towards fully automated machine learning in medical imaging.
MRGen: Diffusion-based Controllable Data Engine for MRI Segmentation towards Unannotated Modalities
Dec 04, 2024



Abstract:Medical image segmentation has recently demonstrated impressive progress with deep neural networks, yet the heterogeneous modalities and scarcity of mask annotations limit the development of segmentation models on unannotated modalities. This paper investigates a new paradigm for leveraging generative models in medical applications: controllably synthesizing data for unannotated modalities, without requiring registered data pairs. Specifically, we make the following contributions in this paper: (i) we collect and curate a large-scale radiology image-text dataset, MedGen-1M, comprising modality labels, attributes, region, and organ information, along with a subset of organ mask annotations, to support research in controllable medical image generation; (ii) we propose a diffusion-based data engine, termed MRGen, which enables generation conditioned on text prompts and masks, synthesizing MR images for diverse modalities lacking mask annotations, to train segmentation models on unannotated modalities; (iii) we conduct extensive experiments across various modalities, illustrating that our data engine can effectively synthesize training samples and extend MRI segmentation towards unannotated modalities.
LoRKD: Low-Rank Knowledge Decomposition for Medical Foundation Models
Sep 29, 2024



Abstract:The widespread adoption of large-scale pre-training techniques has significantly advanced the development of medical foundation models, enabling them to serve as versatile tools across a broad range of medical tasks. However, despite their strong generalization capabilities, medical foundation models pre-trained on large-scale datasets tend to suffer from domain gaps between heterogeneous data, leading to suboptimal performance on specific tasks compared to specialist models, as evidenced by previous studies. In this paper, we explore a new perspective called "Knowledge Decomposition" to improve the performance on specific medical tasks, which deconstructs the foundation model into multiple lightweight expert models, each dedicated to a particular anatomical region, with the aim of enhancing specialization and simultaneously reducing resource consumption. To accomplish the above objective, we propose a novel framework named Low-Rank Knowledge Decomposition (LoRKD), which explicitly separates gradients from different tasks by incorporating low-rank expert modules and efficient knowledge separation convolution. The low-rank expert modules resolve gradient conflicts between heterogeneous data from different anatomical regions, providing strong specialization at lower costs. The efficient knowledge separation convolution significantly improves algorithm efficiency by achieving knowledge separation within a single forward propagation. Extensive experimental results on segmentation and classification tasks demonstrate that our decomposed models not only achieve state-of-the-art performance but also exhibit superior transferability on downstream tasks, even surpassing the original foundation models in task-specific evaluations. The code is available at here.
RadGenome-Chest CT: A Grounded Vision-Language Dataset for Chest CT Analysis
Apr 25, 2024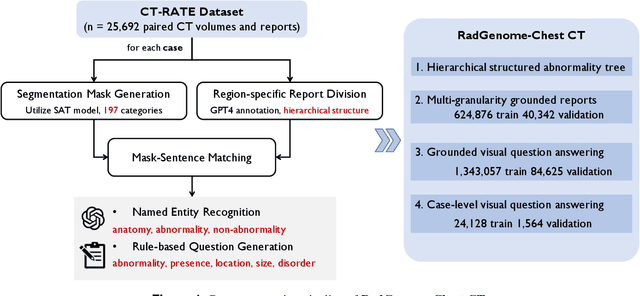


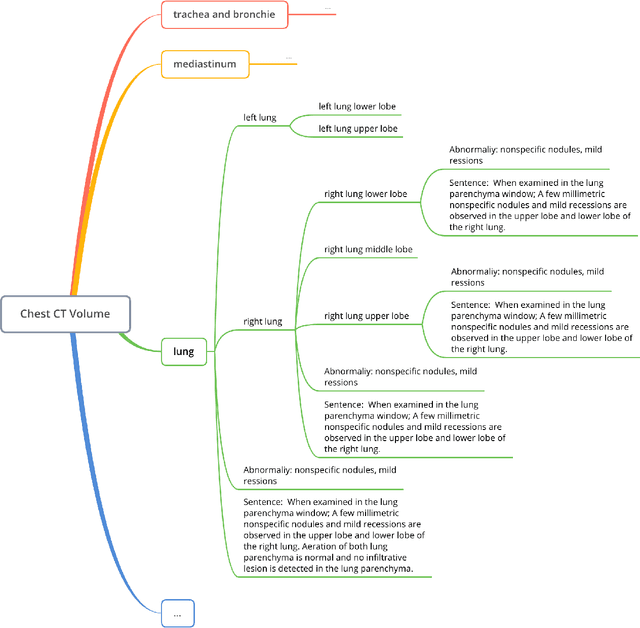
Abstract:Developing generalist foundation model has recently attracted tremendous attention among researchers in the field of AI for Medicine (AI4Medicine). A pivotal insight in developing these models is their reliance on dataset scaling, which emphasizes the requirements on developing open-source medical image datasets that incorporate diverse supervision signals across various imaging modalities. In this paper, we introduce RadGenome-Chest CT, a comprehensive, large-scale, region-guided 3D chest CT interpretation dataset based on CT-RATE. Specifically, we leverage the latest powerful universal segmentation and large language models, to extend the original datasets (over 25,692 non-contrast 3D chest CT volume and reports from 20,000 patients) from the following aspects: (i) organ-level segmentation masks covering 197 categories, which provide intermediate reasoning visual clues for interpretation; (ii) 665 K multi-granularity grounded reports, where each sentence of the report is linked to the corresponding anatomical region of CT volume in the form of a segmentation mask; (iii) 1.3 M grounded VQA pairs, where questions and answers are all linked with reference segmentation masks, enabling models to associate visual evidence with textual explanations. All grounded reports and VQA pairs in the validation set have gone through manual verification to ensure dataset quality. We believe that RadGenome-Chest CT can significantly advance the development of multimodal medical foundation models, by training to generate texts based on given segmentation regions, which is unattainable with previous relevant datasets. We will release all segmentation masks, grounded reports, and VQA pairs to facilitate further research and development in this field.
One Model to Rule them All: Towards Universal Segmentation for Medical Images with Text Prompts
Dec 28, 2023



Abstract:In this study, we focus on building up a model that can Segment Anything in medical scenarios, driven by Text prompts, termed as SAT. Our main contributions are three folds: (i) on data construction, we combine multiple knowledge sources to construct a multi-modal medical knowledge tree; Then we build up a large-scale segmentation dataset for training, by collecting over 11K 3D medical image scans from 31 segmentation datasets with careful standardization on both visual scans and label space; (ii) on model training, we formulate a universal segmentation model, that can be prompted by inputting medical terminologies in text form. We present a knowledge-enhanced representation learning framework, and a series of strategies for effectively training on the combination of a large number of datasets; (iii) on model evaluation, we train a SAT-Nano with only 107M parameters, to segment 31 different segmentation datasets with text prompt, resulting in 362 categories. We thoroughly evaluate the model from three aspects: averaged by body regions, averaged by classes, and average by datasets, demonstrating comparable performance to 36 specialist nnUNets, i.e., we train nnUNet models on each dataset/subset, resulting in 36 nnUNets with around 1000M parameters for the 31 datasets. We will release all the codes, and models used in this report, i.e., SAT-Nano. Moreover, we will offer SAT-Ultra in the near future, which is trained with model of larger size, on more diverse datasets. Webpage URL: https://zhaoziheng.github.io/MedUniSeg.
Can GPT-4V Serve Medical Applications? Case Studies on GPT-4V for Multimodal Medical Diagnosis
Oct 17, 2023Abstract:Driven by the large foundation models, the development of artificial intelligence has witnessed tremendous progress lately, leading to a surge of general interest from the public. In this study, we aim to assess the performance of OpenAI's newest model, GPT-4V(ision), specifically in the realm of multimodal medical diagnosis. Our evaluation encompasses 17 human body systems, including Central Nervous System, Head and Neck, Cardiac, Chest, Hematology, Hepatobiliary, Gastrointestinal, Urogenital, Gynecology, Obstetrics, Breast, Musculoskeletal, Spine, Vascular, Oncology, Trauma, Pediatrics, with images taken from 8 modalities used in daily clinic routine, e.g., X-ray, Computed Tomography (CT), Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), Digital Subtraction Angiography (DSA), Mammography, Ultrasound, and Pathology. We probe the GPT-4V's ability on multiple clinical tasks with or without patent history provided, including imaging modality and anatomy recognition, disease diagnosis, report generation, disease localisation. Our observation shows that, while GPT-4V demonstrates proficiency in distinguishing between medical image modalities and anatomy, it faces significant challenges in disease diagnosis and generating comprehensive reports. These findings underscore that while large multimodal models have made significant advancements in computer vision and natural language processing, it remains far from being used to effectively support real-world medical applications and clinical decision-making. All images used in this report can be found in https://github.com/chaoyi-wu/GPT-4V_Medical_Evaluation.
PMC-VQA: Visual Instruction Tuning for Medical Visual Question Answering
May 24, 2023



Abstract:In this paper, we focus on the problem of Medical Visual Question Answering (MedVQA), which is crucial in efficiently interpreting medical images with vital clinic-relevant information. Firstly, we reframe the problem of MedVQA as a generation task that naturally follows the human-machine interaction, we propose a generative-based model for medical visual understanding by aligning visual information from a pre-trained vision encoder with a large language model. Secondly, we establish a scalable pipeline to construct a large-scale medical visual question-answering dataset, named PMC-VQA, which contains 227k VQA pairs of 149k images that cover various modalities or diseases. Thirdly, we pre-train our proposed model on PMC-VQA and then fine-tune it on multiple public benchmarks, e.g., VQA-RAD and SLAKE, outperforming existing work by a large margin. Additionally, we propose a test set that has undergone manual verification, which is significantly more challenging, even the best models struggle to solve.
PMC-CLIP: Contrastive Language-Image Pre-training using Biomedical Documents
Mar 13, 2023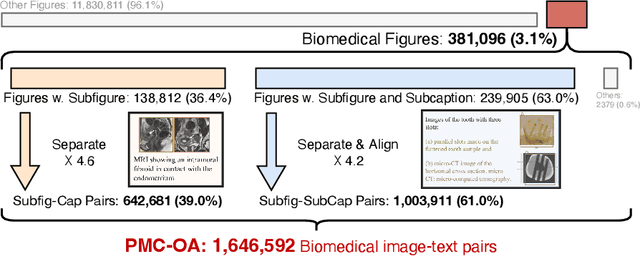
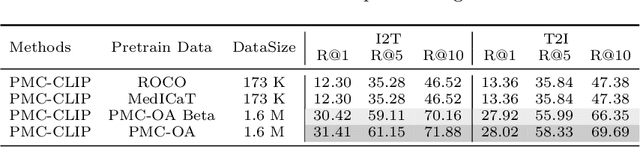
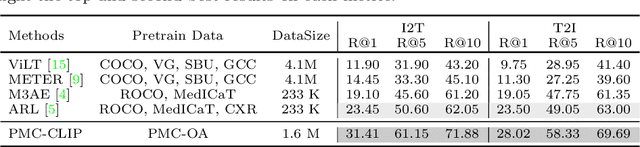
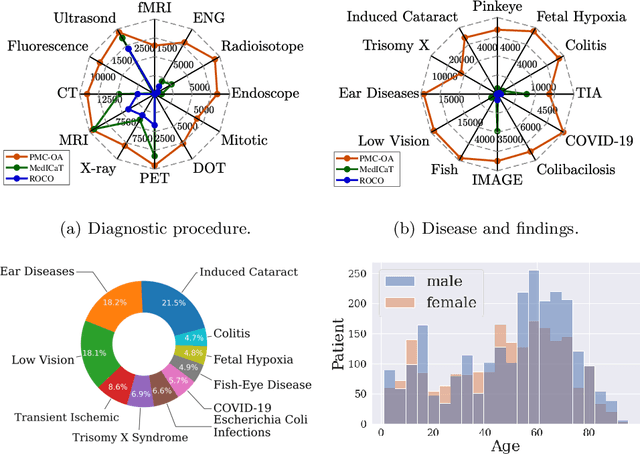
Abstract:Foundation models trained on large-scale dataset gain a recent surge in CV and NLP. In contrast, development in biomedical domain lags far behind due to data scarcity. To address this issue, we build and release PMC-OA, a biomedical dataset with 1.6M image-caption pairs collected from PubMedCentral's OpenAccess subset, which is 8 times larger than before. PMC-OA covers diverse modalities or diseases, with majority of the image-caption samples aligned at finer-grained level, i.e., subfigure and subcaption. While pretraining a CLIP-style model on PMC-OA, our model named PMC-CLIP achieves state-of-the-art results on various downstream tasks, including image-text retrieval on ROCO, MedMNIST image classification, Medical VQA, i.e. +8.1% R@10 on image-text retrieval, +3.9% accuracy on image classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge