Weixiong Lin
SERM: Self-Evolving Relevance Model with Agent-Driven Learning from Massive Query Streams
Jan 14, 2026Abstract:Due to the dynamically evolving nature of real-world query streams, relevance models struggle to generalize to practical search scenarios. A sophisticated solution is self-evolution techniques. However, in large-scale industrial settings with massive query streams, this technique faces two challenges: (1) informative samples are often sparse and difficult to identify, and (2) pseudo-labels generated by the current model could be unreliable. To address these challenges, in this work, we propose a Self-Evolving Relevance Model approach (SERM), which comprises two complementary multi-agent modules: a multi-agent sample miner, designed to detect distributional shifts and identify informative training samples, and a multi-agent relevance annotator, which provides reliable labels through a two-level agreement framework. We evaluate SERM in a large-scale industrial setting, which serves billions of user requests daily. Experimental results demonstrate that SERM can achieve significant performance gains through iterative self-evolution, as validated by extensive offline multilingual evaluations and online testing.
Squeeze Out Tokens from Sample for Finer-Grained Data Governance
Mar 18, 2025Abstract:Widely observed data scaling laws, in which error falls off as a power of the training size, demonstrate the diminishing returns of unselective data expansion. Hence, data governance is proposed to downsize datasets through pruning non-informative samples. Yet, isolating the impact of a specific sample on overall model performance is challenging, due to the vast computation required for tryout all sample combinations. Current data governors circumvent this complexity by estimating sample contributions through heuristic-derived scalar scores, thereby discarding low-value ones. Despite thorough sample sieving, retained samples contain substantial undesired tokens intrinsically, underscoring the potential for further compression and purification. In this work, we upgrade data governance from a 'sieving' approach to a 'juicing' one. Instead of scanning for least-flawed samples, our dual-branch DataJuicer applies finer-grained intra-sample governance. It squeezes out informative tokens and boosts image-text alignments. Specifically, the vision branch retains salient image patches and extracts relevant object classes, while the text branch incorporates these classes to enhance captions. Consequently, DataJuicer yields more refined datasets through finer-grained governance. Extensive experiments across datasets demonstrate that DataJuicer significantly outperforms existing DataSieve in image-text retrieval, classification, and dense visual reasoning.
Contrast-Unity for Partially-Supervised Temporal Sentence Grounding
Feb 18, 2025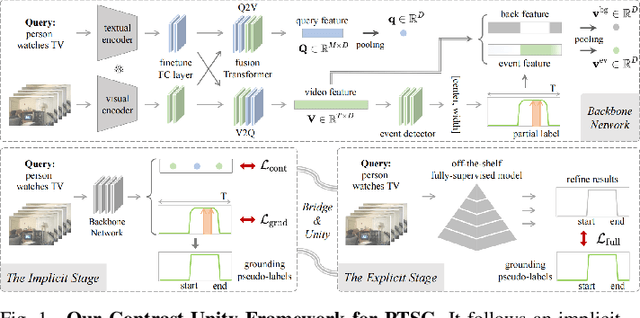
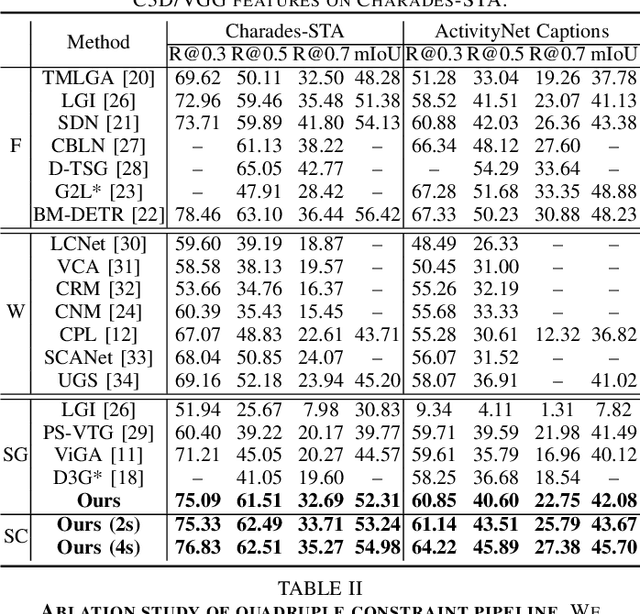
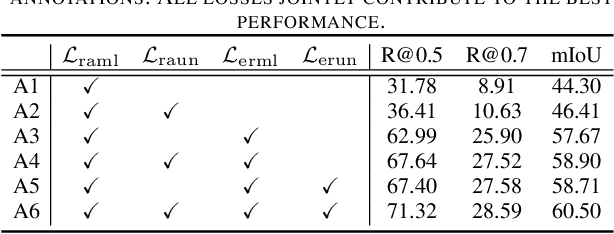
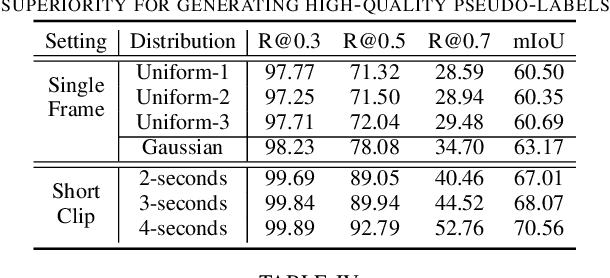
Abstract:Temporal sentence grounding aims to detect event timestamps described by the natural language query from given untrimmed videos. The existing fully-supervised setting achieves great results but requires expensive annotation costs; while the weakly-supervised setting adopts cheap labels but performs poorly. To pursue high performance with less annotation costs, this paper introduces an intermediate partially-supervised setting, i.e., only short-clip is available during training. To make full use of partial labels, we specially design one contrast-unity framework, with the two-stage goal of implicit-explicit progressive grounding. In the implicit stage, we align event-query representations at fine granularity using comprehensive quadruple contrastive learning: event-query gather, event-background separation, intra-cluster compactness and inter-cluster separability. Then, high-quality representations bring acceptable grounding pseudo-labels. In the explicit stage, to explicitly optimize grounding objectives, we train one fully-supervised model using obtained pseudo-labels for grounding refinement and denoising. Extensive experiments and thoroughly ablations on Charades-STA and ActivityNet Captions demonstrate the significance of partial supervision, as well as our superior performance.
Advancing Myopia To Holism: Fully Contrastive Language-Image Pre-training
Nov 30, 2024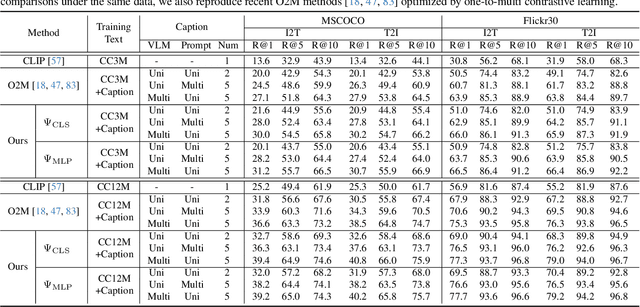
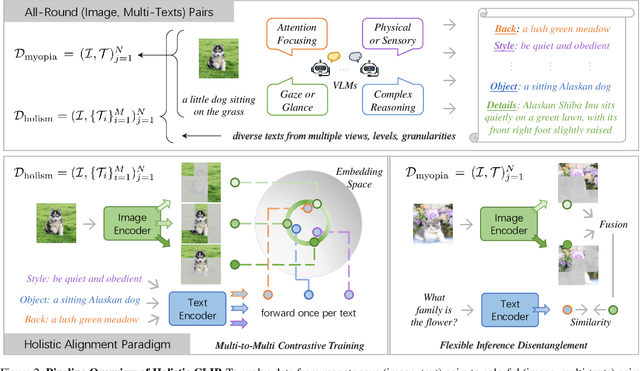
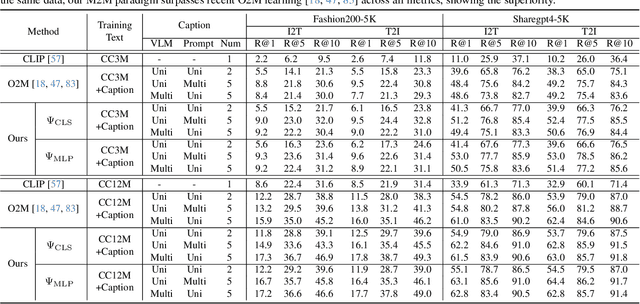
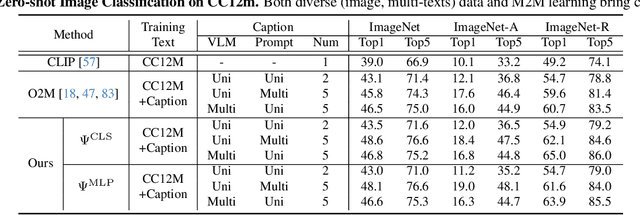
Abstract:In rapidly evolving field of vision-language models (VLMs), contrastive language-image pre-training (CLIP) has made significant strides, becoming foundation for various downstream tasks. However, relying on one-to-one (image, text) contrastive paradigm to learn alignment from large-scale messy web data, CLIP faces a serious myopic dilemma, resulting in biases towards monotonous short texts and shallow visual expressivity. To overcome these issues, this paper advances CLIP into one novel holistic paradigm, by updating both diverse data and alignment optimization. To obtain colorful data with low cost, we use image-to-text captioning to generate multi-texts for each image, from multiple perspectives, granularities, and hierarchies. Two gadgets are proposed to encourage textual diversity. To match such (image, multi-texts) pairs, we modify the CLIP image encoder into multi-branch, and propose multi-to-multi contrastive optimization for image-text part-to-part matching. As a result, diverse visual embeddings are learned for each image, bringing good interpretability and generalization. Extensive experiments and ablations across over ten benchmarks indicate that our holistic CLIP significantly outperforms existing myopic CLIP, including image-text retrieval, open-vocabulary classification, and dense visual tasks.
Prompt Tuning with Diffusion for Few-Shot Pre-trained Policy Generalization
Nov 02, 2024



Abstract:Offline reinforcement learning (RL) methods harness previous experiences to derive an optimal policy, forming the foundation for pre-trained large-scale models (PLMs). When encountering tasks not seen before, PLMs often utilize several expert trajectories as prompts to expedite their adaptation to new requirements. Though a range of prompt-tuning methods have been proposed to enhance the quality of prompts, these methods often face optimization restrictions due to prompt initialization, which can significantly constrain the exploration domain and potentially lead to suboptimal solutions. To eliminate the reliance on the initial prompt, we shift our perspective towards the generative model, framing the prompt-tuning process as a form of conditional generative modeling, where prompts are generated from random noise. Our innovation, the Prompt Diffuser, leverages a conditional diffusion model to produce prompts of exceptional quality. Central to our framework is the approach to trajectory reconstruction and the meticulous integration of downstream task guidance during the training phase. Further experimental results underscore the potency of the Prompt Diffuser as a robust and effective tool for the prompt-tuning process, demonstrating strong performance in the meta-RL tasks.
Towards Building Multilingual Language Model for Medicine
Feb 26, 2024



Abstract:In this paper, we aim to develop an open-source, multilingual language model for medicine, that the benefits a wider, linguistically diverse audience from different regions. In general, we present the contribution from the following aspects: first, for multilingual medical-specific adaptation, we construct a new multilingual medical corpus, that contains approximately 25.5B tokens encompassing 6 main languages, termed as MMedC, that enables auto-regressive training for existing general LLMs. second, to monitor the development of multilingual LLMs in medicine, we propose a new multilingual medical multi-choice question-answering benchmark with rationale, termed as MMedBench; third, we have assessed a number of popular, opensource large language models (LLMs) on our benchmark, along with those further auto-regressive trained on MMedC, as a result, our final model, termed as MMedLM 2, with only 7B parameters, achieves superior performance compared to all other open-source models, even rivaling GPT-4 on MMedBench. We will make the resources publicly available, including code, model weights, and datasets.
Can GPT-4V Serve Medical Applications? Case Studies on GPT-4V for Multimodal Medical Diagnosis
Oct 17, 2023Abstract:Driven by the large foundation models, the development of artificial intelligence has witnessed tremendous progress lately, leading to a surge of general interest from the public. In this study, we aim to assess the performance of OpenAI's newest model, GPT-4V(ision), specifically in the realm of multimodal medical diagnosis. Our evaluation encompasses 17 human body systems, including Central Nervous System, Head and Neck, Cardiac, Chest, Hematology, Hepatobiliary, Gastrointestinal, Urogenital, Gynecology, Obstetrics, Breast, Musculoskeletal, Spine, Vascular, Oncology, Trauma, Pediatrics, with images taken from 8 modalities used in daily clinic routine, e.g., X-ray, Computed Tomography (CT), Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), Digital Subtraction Angiography (DSA), Mammography, Ultrasound, and Pathology. We probe the GPT-4V's ability on multiple clinical tasks with or without patent history provided, including imaging modality and anatomy recognition, disease diagnosis, report generation, disease localisation. Our observation shows that, while GPT-4V demonstrates proficiency in distinguishing between medical image modalities and anatomy, it faces significant challenges in disease diagnosis and generating comprehensive reports. These findings underscore that while large multimodal models have made significant advancements in computer vision and natural language processing, it remains far from being used to effectively support real-world medical applications and clinical decision-making. All images used in this report can be found in https://github.com/chaoyi-wu/GPT-4V_Medical_Evaluation.
PMC-VQA: Visual Instruction Tuning for Medical Visual Question Answering
May 24, 2023



Abstract:In this paper, we focus on the problem of Medical Visual Question Answering (MedVQA), which is crucial in efficiently interpreting medical images with vital clinic-relevant information. Firstly, we reframe the problem of MedVQA as a generation task that naturally follows the human-machine interaction, we propose a generative-based model for medical visual understanding by aligning visual information from a pre-trained vision encoder with a large language model. Secondly, we establish a scalable pipeline to construct a large-scale medical visual question-answering dataset, named PMC-VQA, which contains 227k VQA pairs of 149k images that cover various modalities or diseases. Thirdly, we pre-train our proposed model on PMC-VQA and then fine-tune it on multiple public benchmarks, e.g., VQA-RAD and SLAKE, outperforming existing work by a large margin. Additionally, we propose a test set that has undergone manual verification, which is significantly more challenging, even the best models struggle to solve.
PMC-CLIP: Contrastive Language-Image Pre-training using Biomedical Documents
Mar 13, 2023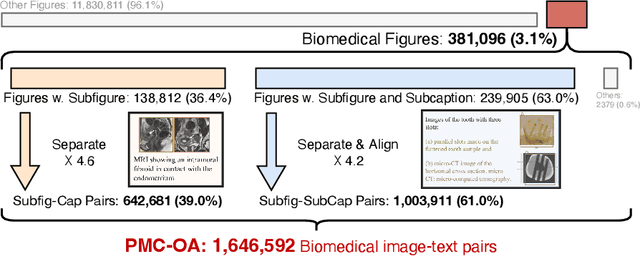
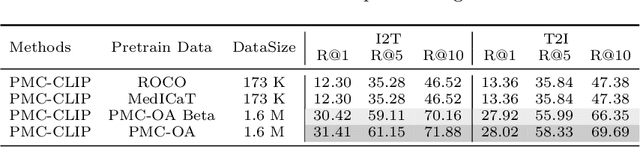
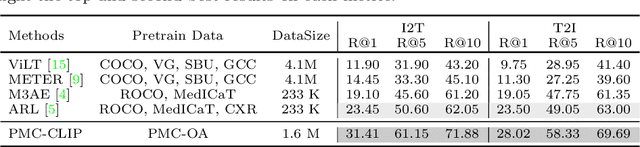
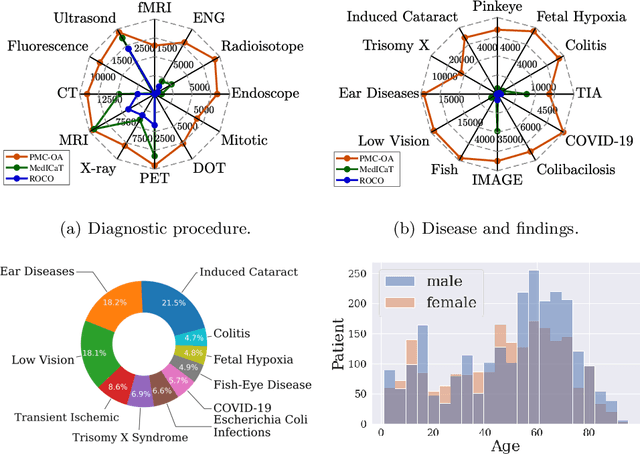
Abstract:Foundation models trained on large-scale dataset gain a recent surge in CV and NLP. In contrast, development in biomedical domain lags far behind due to data scarcity. To address this issue, we build and release PMC-OA, a biomedical dataset with 1.6M image-caption pairs collected from PubMedCentral's OpenAccess subset, which is 8 times larger than before. PMC-OA covers diverse modalities or diseases, with majority of the image-caption samples aligned at finer-grained level, i.e., subfigure and subcaption. While pretraining a CLIP-style model on PMC-OA, our model named PMC-CLIP achieves state-of-the-art results on various downstream tasks, including image-text retrieval on ROCO, MedMNIST image classification, Medical VQA, i.e. +8.1% R@10 on image-text retrieval, +3.9% accuracy on image classification.
GnetSeg: Semantic Segmentation Model Optimized on a 224mW CNN Accelerator Chip at the Speed of 318FPS
Jan 09, 2021

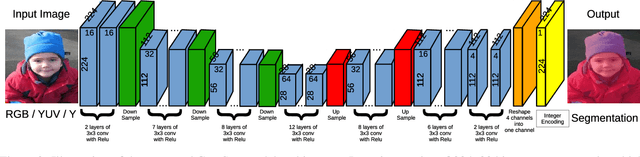
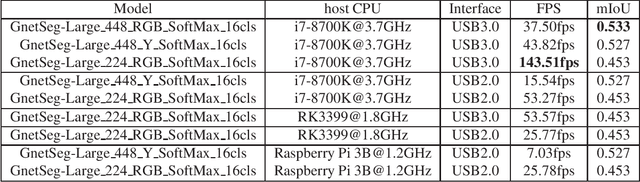
Abstract:Semantic segmentation is the task to cluster pixels on an image belonging to the same class. It is widely used in the real-world applications including autonomous driving, medical imaging analysis, industrial inspection, smartphone camera for person segmentation and so on. Accelerating the semantic segmentation models on the mobile and edge devices are practical needs for the industry. Recent years have witnessed the wide availability of CNN (Convolutional Neural Networks) accelerators. They have the advantages on power efficiency, inference speed, which are ideal for accelerating the semantic segmentation models on the edge devices. However, the CNN accelerator chips also have the limitations on flexibility and memory. In addition, the CPU load is very critical because the CNN accelerator chip works as a co-processor with a host CPU. In this paper, we optimize the semantic segmentation model in order to fully utilize the limited memory and the supported operators on the CNN accelerator chips, and at the same time reduce the CPU load of the CNN model to zero. The resulting model is called GnetSeg. Furthermore, we propose the integer encoding for the mask of the GnetSeg model, which minimizes the latency of data transfer between the CNN accelerator and the host CPU. The experimental result shows that the model running on the 224mW chip achieves the speed of 318FPS with excellent accuracy for applications such as person segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge