Monique Haak
Exploring Test Time Adaptation for Subcortical Segmentation of the Fetal Brain in 3D Ultrasound
Feb 12, 2025Abstract:Monitoring the growth of subcortical regions of the fetal brain in ultrasound (US) images can help identify the presence of abnormal development. Manually segmenting these regions is a challenging task, but recent work has shown that it can be automated using deep learning. However, applying pretrained models to unseen freehand US volumes often leads to a degradation of performance due to the vast differences in acquisition and alignment. In this work, we first demonstrate that test time adaptation (TTA) can be used to improve model performance in the presence of both real and simulated domain shifts. We further propose a novel TTA method by incorporating a normative atlas as a prior for anatomy. In the presence of various types of domain shifts, we benchmark the performance of different TTA methods and demonstrate the improvements brought by our proposed approach, which may further facilitate automated monitoring of fetal brain development. Our code is available at https://github.com/joshuaomolegan/TTA-for-3D-Fetal-Subcortical-Segmentation.
Adaptive 3D Localization of 2D Freehand Ultrasound Brain Images
Sep 12, 2022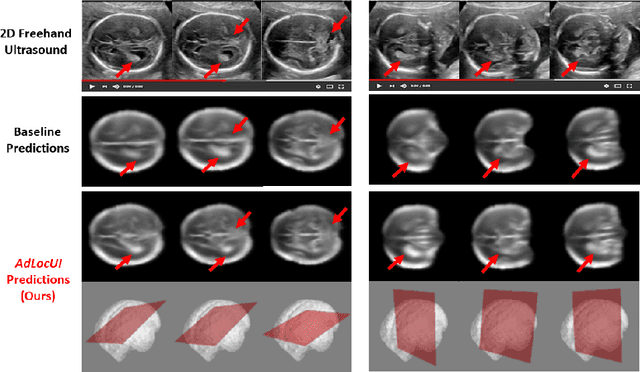
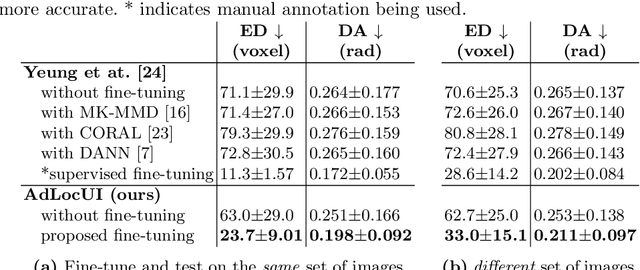
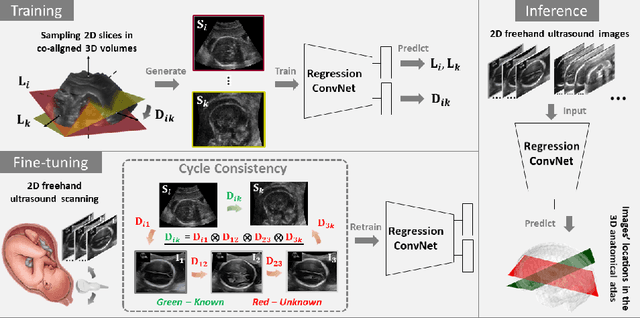
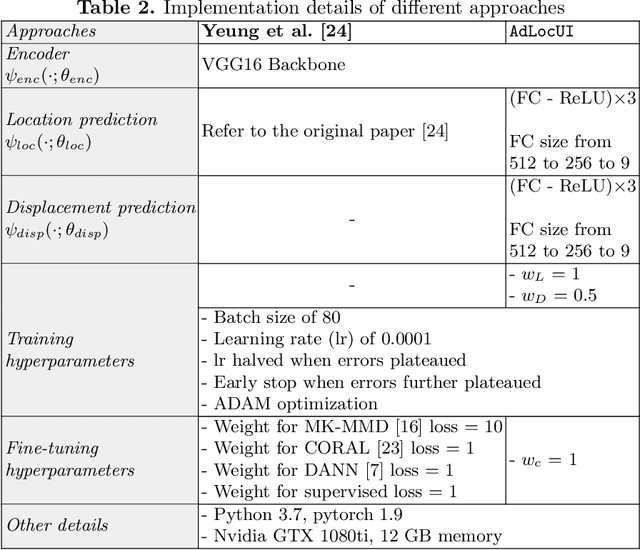
Abstract:Two-dimensional (2D) freehand ultrasound is the mainstay in prenatal care and fetal growth monitoring. The task of matching corresponding cross-sectional planes in the 3D anatomy for a given 2D ultrasound brain scan is essential in freehand scanning, but challenging. We propose AdLocUI, a framework that Adaptively Localizes 2D Ultrasound Images in the 3D anatomical atlas without using any external tracking sensor.. We first train a convolutional neural network with 2D slices sampled from co-aligned 3D ultrasound volumes to predict their locations in the 3D anatomical atlas. Next, we fine-tune it with 2D freehand ultrasound images using a novel unsupervised cycle consistency, which utilizes the fact that the overall displacement of a sequence of images in the 3D anatomical atlas is equal to the displacement from the first image to the last in that sequence. We demonstrate that AdLocUI can adapt to three different ultrasound datasets, acquired with different machines and protocols, and achieves significantly better localization accuracy than the baselines. AdLocUI can be used for sensorless 2D freehand ultrasound guidance by the bedside. The source code is available at https://github.com/pakheiyeung/AdLocUI.
ImplicitVol: Sensorless 3D Ultrasound Reconstruction with Deep Implicit Representation
Sep 24, 2021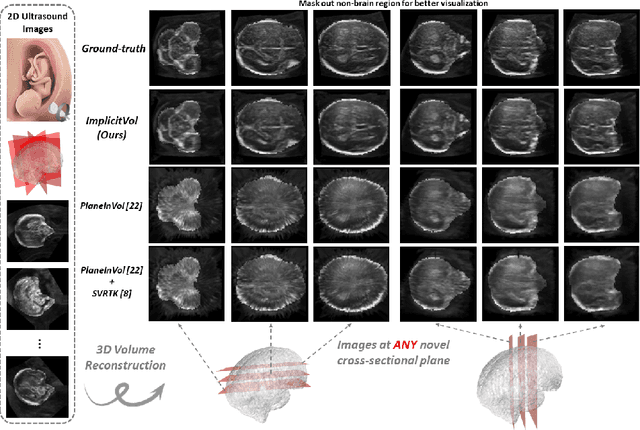
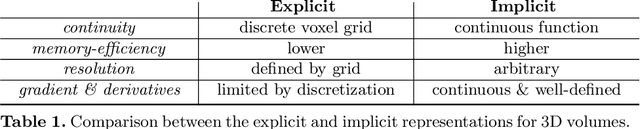
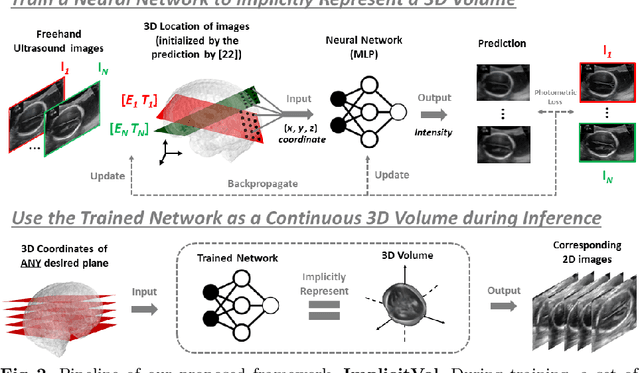
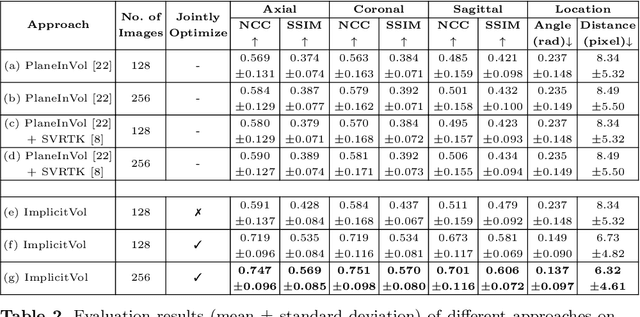
Abstract:The objective of this work is to achieve sensorless reconstruction of a 3D volume from a set of 2D freehand ultrasound images with deep implicit representation. In contrast to the conventional way that represents a 3D volume as a discrete voxel grid, we do so by parameterizing it as the zero level-set of a continuous function, i.e. implicitly representing the 3D volume as a mapping from the spatial coordinates to the corresponding intensity values. Our proposed model, termed as ImplicitVol, takes a set of 2D scans and their estimated locations in 3D as input, jointly re?fing the estimated 3D locations and learning a full reconstruction of the 3D volume. When testing on real 2D ultrasound images, novel cross-sectional views that are sampled from ImplicitVol show significantly better visual quality than those sampled from existing reconstruction approaches, outperforming them by over 30% (NCC and SSIM), between the output and ground-truth on the 3D volume testing data. The code will be made publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge