Madeleine K. Wyburd
rNCA: Self-Repairing Segmentation Masks
Dec 15, 2025Abstract:Accurately predicting topologically correct masks remains a difficult task for general segmentation models, which often produce fragmented or disconnected outputs. Fixing these artifacts typically requires hand-crafted refinement rules or architectures specialized to a particular task. Here, we show that Neural Cellular Automata (NCA) can be directly re-purposed as an effective refinement mechanism, using local, iterative updates guided by image context to repair segmentation masks. By training on imperfect masks and ground truths, the automaton learns the structural properties of the target shape while relying solely on local information. When applied to coarse, globally predicted masks, the learned dynamics progressively reconnect broken regions, prune loose fragments and converge towards stable, topologically consistent results. We show how refinement NCA (rNCA) can be easily applied to repair common topological errors produced by different base segmentation models and tasks: for fragmented retinal vessels, it yields 2-3% gains in Dice/clDice and improves Betti errors, reducing $β_0$ errors by 60% and $β_1$ by 20%; for myocardium, it repairs 61.5% of broken cases in a zero-shot setting while lowering ASSD and HD by 19% and 16%, respectively. This showcases NCA as effective and broadly applicable refiners.
Automated Fetal Biometry Assessment with Deep Ensembles using Sparse-Sampling of 2D Intrapartum Ultrasound Images
May 20, 2025



Abstract:The International Society of Ultrasound advocates Intrapartum Ultrasound (US) Imaging in Obstetrics and Gynecology (ISUOG) to monitor labour progression through changes in fetal head position. Two reliable ultrasound-derived parameters that are used to predict outcomes of instrumental vaginal delivery are the angle of progression (AoP) and head-symphysis distance (HSD). In this work, as part of the Intrapartum Ultrasounds Grand Challenge (IUGC) 2024, we propose an automated fetal biometry measurement pipeline to reduce intra- and inter-observer variability and improve measurement reliability. Our pipeline consists of three key tasks: (i) classification of standard planes (SP) from US videos, (ii) segmentation of fetal head and pubic symphysis from the detected SPs, and (iii) computation of the AoP and HSD from the segmented regions. We perform sparse sampling to mitigate class imbalances and reduce spurious correlations in task (i), and utilize ensemble-based deep learning methods for task (i) and (ii) to enhance generalizability under different US acquisition settings. Finally, to promote robustness in task iii) with respect to the structural fidelity of measurements, we retain the largest connected components and apply ellipse fitting to the segmentations. Our solution achieved ACC: 0.9452, F1: 0.9225, AUC: 0.983, MCC: 0.8361, DSC: 0.918, HD: 19.73, ASD: 5.71, $\Delta_{AoP}$: 8.90 and $\Delta_{HSD}$: 14.35 across an unseen hold-out set of 4 patients and 224 US frames. The results from the proposed automated pipeline can improve the understanding of labour arrest causes and guide the development of clinical risk stratification tools for efficient and effective prenatal care.
Exploring Test Time Adaptation for Subcortical Segmentation of the Fetal Brain in 3D Ultrasound
Feb 12, 2025Abstract:Monitoring the growth of subcortical regions of the fetal brain in ultrasound (US) images can help identify the presence of abnormal development. Manually segmenting these regions is a challenging task, but recent work has shown that it can be automated using deep learning. However, applying pretrained models to unseen freehand US volumes often leads to a degradation of performance due to the vast differences in acquisition and alignment. In this work, we first demonstrate that test time adaptation (TTA) can be used to improve model performance in the presence of both real and simulated domain shifts. We further propose a novel TTA method by incorporating a normative atlas as a prior for anatomy. In the presence of various types of domain shifts, we benchmark the performance of different TTA methods and demonstrate the improvements brought by our proposed approach, which may further facilitate automated monitoring of fetal brain development. Our code is available at https://github.com/joshuaomolegan/TTA-for-3D-Fetal-Subcortical-Segmentation.
RapidVol: Rapid Reconstruction of 3D Ultrasound Volumes from Sensorless 2D Scans
Apr 16, 2024



Abstract:Two-dimensional (2D) freehand ultrasonography is one of the most commonly used medical imaging modalities, particularly in obstetrics and gynaecology. However, it only captures 2D cross-sectional views of inherently 3D anatomies, losing valuable contextual information. As an alternative to requiring costly and complex 3D ultrasound scanners, 3D volumes can be constructed from 2D scans using machine learning. However this usually requires long computational time. Here, we propose RapidVol: a neural representation framework to speed up slice-to-volume ultrasound reconstruction. We use tensor-rank decomposition, to decompose the typical 3D volume into sets of tri-planes, and store those instead, as well as a small neural network. A set of 2D ultrasound scans, with their ground truth (or estimated) 3D position and orientation (pose) is all that is required to form a complete 3D reconstruction. Reconstructions are formed from real fetal brain scans, and then evaluated by requesting novel cross-sectional views. When compared to prior approaches based on fully implicit representation (e.g. neural radiance fields), our method is over 3x quicker, 46% more accurate, and if given inaccurate poses is more robust. Further speed-up is also possible by reconstructing from a structural prior rather than from scratch.
TEDS-Net: Enforcing Diffeomorphisms in Spatial Transformers to Guarantee Topology Preservation in Segmentations
Jul 28, 2021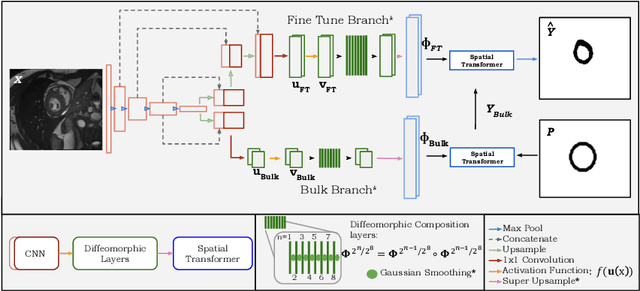
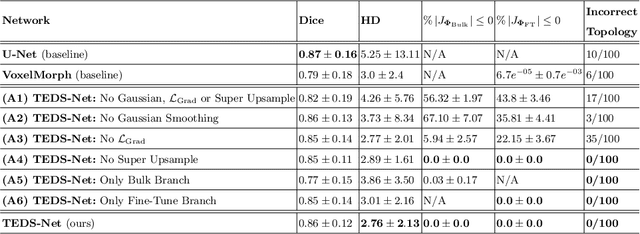


Abstract:Accurate topology is key when performing meaningful anatomical segmentations, however, it is often overlooked in traditional deep learning methods. In this work we propose TEDS-Net: a novel segmentation method that guarantees accurate topology. Our method is built upon a continuous diffeomorphic framework, which enforces topology preservation. However, in practice, diffeomorphic fields are represented using a finite number of parameters and sampled using methods such as linear interpolation, violating the theoretical guarantees. We therefore introduce additional modifications to more strictly enforce it. Our network learns how to warp a binary prior, with the desired topological characteristics, to complete the segmentation task. We tested our method on myocardium segmentation from an open-source 2D heart dataset. TEDS-Net preserved topology in 100% of the cases, compared to 90% from the U-Net, without sacrificing on Hausdorff Distance or Dice performance. Code will be made available at: www.github.com/mwyburd/TEDS-Net
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge