Linde S. Hesse
Prototype Learning for Explainable Regression
Jun 16, 2023Abstract:The lack of explainability limits the adoption of deep learning models in clinical practice. While methods exist to improve the understanding of such models, these are mainly saliency-based and developed for classification, despite many important tasks in medical imaging being continuous regression problems. Therefore, in this work, we present ExPeRT: an explainable prototype-based model specifically designed for regression tasks. Our proposed model makes a sample prediction from the distances to a set of learned prototypes in latent space, using a weighted mean of prototype labels. The distances in latent space are regularized to be relative to label differences, and each of the prototypes can be visualized as a sample from the training set. The image-level distances are further constructed from patch-level distances, in which the patches of both images are structurally matched using optimal transport. We demonstrate our proposed model on the task of brain age prediction on two image datasets: adult MR and fetal ultrasound. Our approach achieved state-of-the-art prediction performance while providing insight in the model's reasoning process.
INSightR-Net: Interpretable Neural Network for Regression using Similarity-based Comparisons to Prototypical Examples
Jul 31, 2022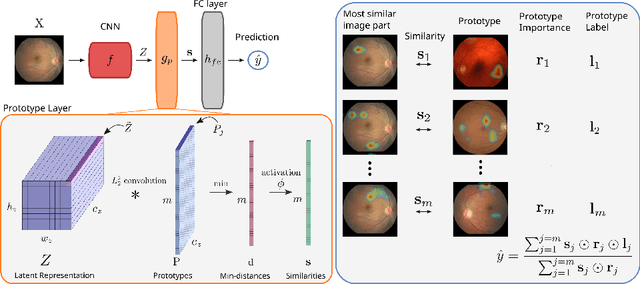
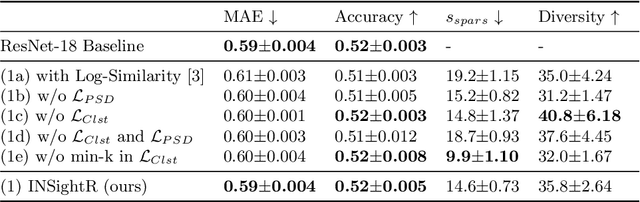
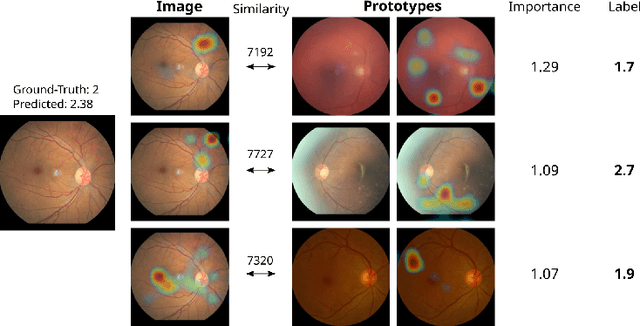
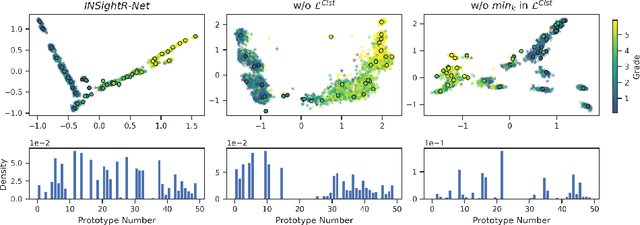
Abstract:Convolutional neural networks (CNNs) have shown exceptional performance for a range of medical imaging tasks. However, conventional CNNs are not able to explain their reasoning process, therefore limiting their adoption in clinical practice. In this work, we propose an inherently interpretable CNN for regression using similarity-based comparisons (INSightR-Net) and demonstrate our methods on the task of diabetic retinopathy grading. A prototype layer incorporated into the architecture enables visualization of the areas in the image that are most similar to learned prototypes. The final prediction is then intuitively modeled as a mean of prototype labels, weighted by the similarities. We achieved competitive prediction performance with our INSightR-Net compared to a ResNet baseline, showing that it is not necessary to compromise performance for interpretability. Furthermore, we quantified the quality of our explanations using sparsity and diversity, two concepts considered important for a good explanation, and demonstrated the effect of several parameters on the latent space embeddings.
Primary Tumor Origin Classification of Lung Nodules in Spectral CT using Transfer Learning
Jun 30, 2020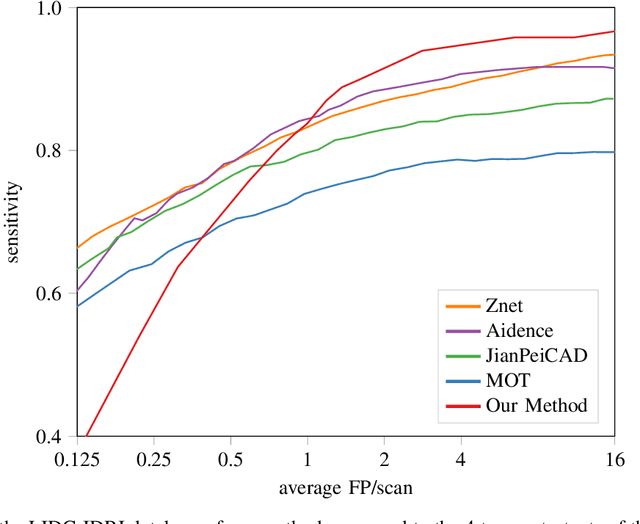
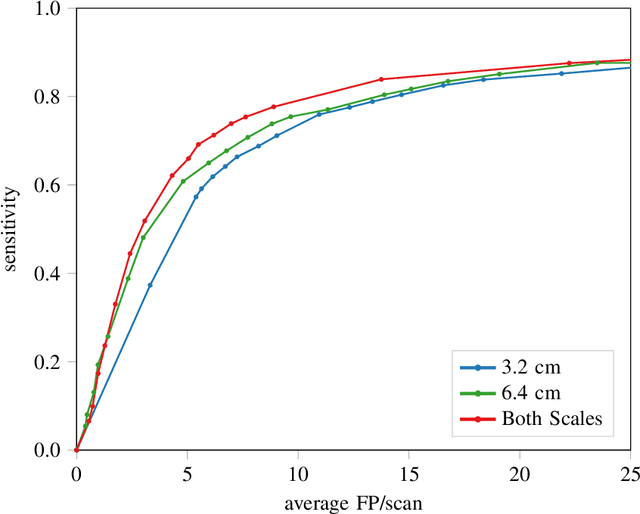
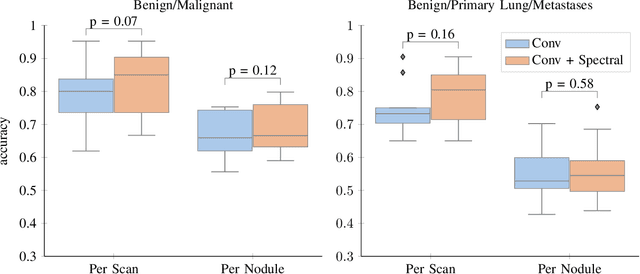
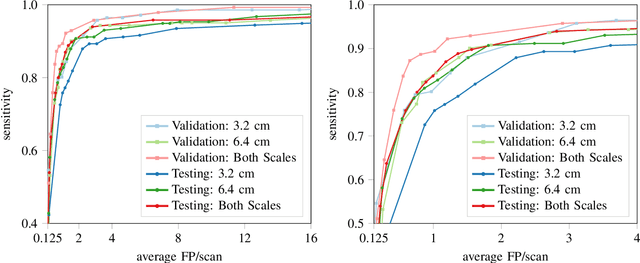
Abstract:Early detection of lung cancer has been proven to decrease mortality significantly. A recent development in computed tomography (CT), spectral CT, can potentially improve diagnostic accuracy, as it yields more information per scan than regular CT. However, the shear workload involved with analyzing a large number of scans drives the need for automated diagnosis methods. Therefore, we propose a detection and classification system for lung nodules in CT scans. Furthermore, we want to observe whether spectral images can increase classifier performance. For the detection of nodules we trained a VGG-like 3D convolutional neural net (CNN). To obtain a primary tumor classifier for our dataset we pre-trained a 3D CNN with similar architecture on nodule malignancies of a large publicly available dataset, the LIDC-IDRI dataset. Subsequently we used this pre-trained network as feature extractor for the nodules in our dataset. The resulting feature vectors were classified into two (benign/malignant) and three (benign/primary lung cancer/metastases) classes using support vector machine (SVM). This classification was performed both on nodule- and scan-level. We obtained state-of-the art performance for detection and malignancy regression on the LIDC-IDRI database. Classification performance on our own dataset was higher for scan- than for nodule-level predictions. For the three-class scan-level classification we obtained an accuracy of 78\%. Spectral features did increase classifier performance, but not significantly. Our work suggests that a pre-trained feature extractor can be used as primary tumor origin classifier for lung nodules, eliminating the need for elaborate fine-tuning of a new network and large datasets. Code is available at \url{https://github.com/tueimage/lung-nodule-msc-2018}.
Intensity augmentation for domain transfer of whole breast segmentation in MRI
Sep 05, 2019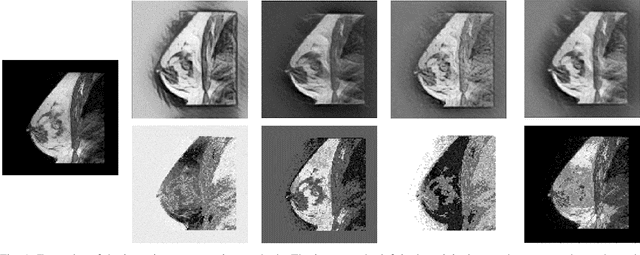
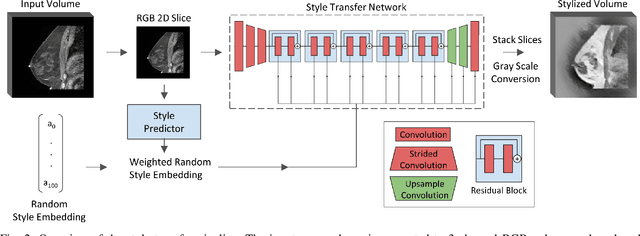
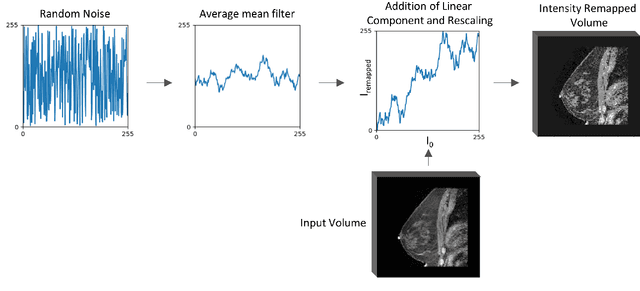
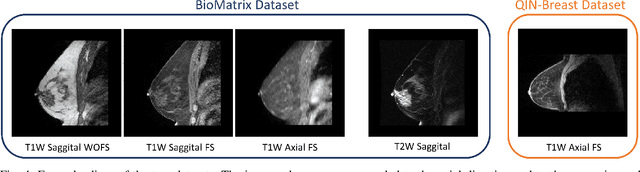
Abstract:The segmentation of the breast from the chest wall is an important first step in the analysis of breast magnetic resonance images. 3D U-nets have been shown to obtain high segmentation accuracy and appear to generalize well when trained on one scanner type and tested on another scanner, provided that a very similar T1-weighted MR protocol is used. There has, however, been little work addressing the problem of domain adaptation when image intensities or patient orientation differ markedly between the training set and an unseen test set. To overcome the domain shift we propose to apply extensive intensity augmentation in addition to geometric augmentation during training. We explored both style transfer and a novel intensity remapping approach as intensity augmentation strategies. For our experiments, we trained a 3D U-net on T1-weighted scans and tested on T2-weighted scans. By applying intensity augmentation we increased segmentation performance from a DSC of 0.71 to 0.90. This performance is very close to the baseline performance of training and testing on T2-weighted scans (0.92). Furthermore, we applied our network to an independent test set made up of publicly available scans acquired using a T1-weighted TWIST sequence and a different coil configuration. On this dataset we obtained a performance of 0.89, close to the inter-observer variability of the ground truth segmentations (0.92). Our results show that using intensity augmentation in addition to geometric augmentation is a suitable method to overcome the intensity domain shift and we expect it to be useful for a wide range of segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge