Aris T. Papageorghiou
Orientation-Robust Latent Motion Trajectory Learning for Annotation-free Cardiac Phase Detection in Fetal Echocardiography
Feb 06, 2026Abstract:Fetal echocardiography is essential for detecting congenital heart disease (CHD), facilitating pregnancy management, optimized delivery planning, and timely postnatal interventions. Among standard imaging planes, the four-chamber (4CH) view provides comprehensive information for CHD diagnosis, where clinicians carefully inspect the end-diastolic (ED) and end-systolic (ES) phases to evaluate cardiac structure and motion. Automated detection of these cardiac phases is thus a critical component toward fully automated CHD analysis. Yet, in the absence of fetal electrocardiography (ECG), manual identification of ED and ES frames remains a labor-intensive bottleneck. We present ORBIT (Orientation-Robust Beat Inference from Trajectories), a self-supervised framework that identifies cardiac phases without manual annotations under various fetal heart orientation. ORBIT employs registration as self-supervision task and learns a latent motion trajectory of cardiac deformation, whose turning points capture transitions between cardiac relaxation and contraction, enabling accurate and orientation-robust localization of ED and ES frames across diverse fetal positions. Trained exclusively on normal fetal echocardiography videos, ORBIT achieves consistent performance on both normal (MAE = 1.9 frames for ED and 1.6 for ES) and CHD cases (MAE = 2.4 frames for ED and 2.1 for ES), outperforming existing annotation-free approaches constrained by fixed orientation assumptions. These results highlight the potential of ORBIT to facilitate robust cardiac phase detection directly from 4CH fetal echocardiography.
Learning to learn skill assessment for fetal ultrasound scanning
Dec 30, 2025Abstract:Traditionally, ultrasound skill assessment has relied on expert supervision and feedback, a process known for its subjectivity and time-intensive nature. Previous works on quantitative and automated skill assessment have predominantly employed supervised learning methods, often limiting the analysis to predetermined or assumed factors considered influential in determining skill levels. In this work, we propose a novel bi-level optimisation framework that assesses fetal ultrasound skills by how well a task is performed on the acquired fetal ultrasound images, without using manually predefined skill ratings. The framework consists of a clinical task predictor and a skill predictor, which are optimised jointly by refining the two networks simultaneously. We validate the proposed method on real-world clinical ultrasound videos of scanning the fetal head. The results demonstrate the feasibility of predicting ultrasound skills by the proposed framework, which quantifies optimised task performance as a skill indicator.
Self-supervised Learning of Echocardiographic Video Representations via Online Cluster Distillation
Jun 13, 2025Abstract:Self-supervised learning (SSL) has achieved major advances in natural images and video understanding, but challenges remain in domains like echocardiography (heart ultrasound) due to subtle anatomical structures, complex temporal dynamics, and the current lack of domain-specific pre-trained models. Existing SSL approaches such as contrastive, masked modeling, and clustering-based methods struggle with high intersample similarity, sensitivity to low PSNR inputs common in ultrasound, or aggressive augmentations that distort clinically relevant features. We present DISCOVR (Distilled Image Supervision for Cross Modal Video Representation), a self-supervised dual branch framework for cardiac ultrasound video representation learning. DISCOVR combines a clustering-based video encoder that models temporal dynamics with an online image encoder that extracts fine-grained spatial semantics. These branches are connected through a semantic cluster distillation loss that transfers anatomical knowledge from the evolving image encoder to the video encoder, enabling temporally coherent representations enriched with fine-grained semantic understanding. Evaluated on six echocardiography datasets spanning fetal, pediatric, and adult populations, DISCOVR outperforms both specialized video anomaly detection methods and state-of-the-art video-SSL baselines in zero-shot and linear probing setups, and achieves superior segmentation transfer.
Pose-GuideNet: Automatic Scanning Guidance for Fetal Head Ultrasound from Pose Estimation
Aug 19, 2024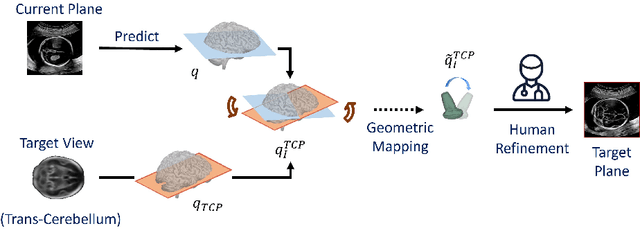
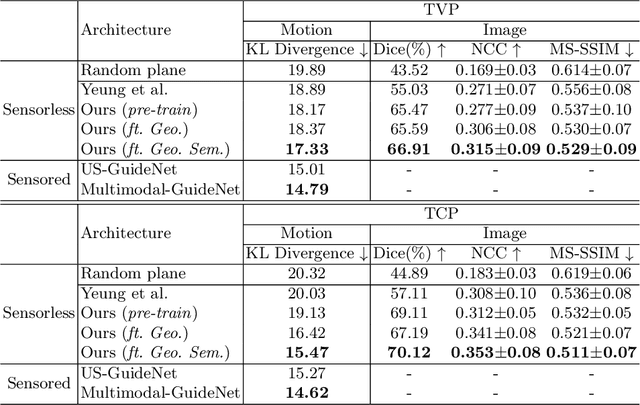
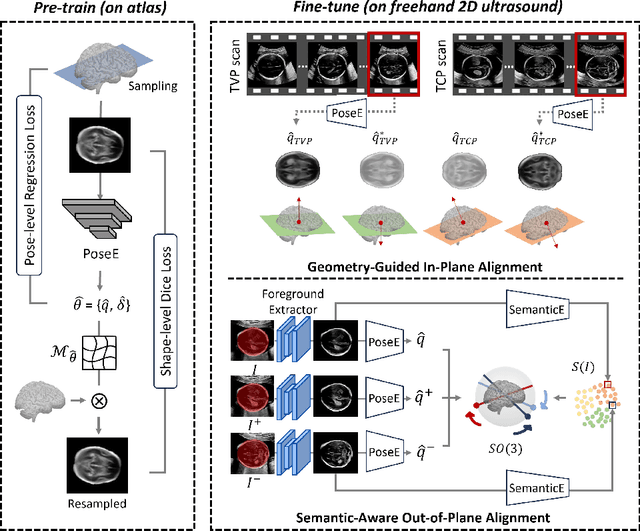
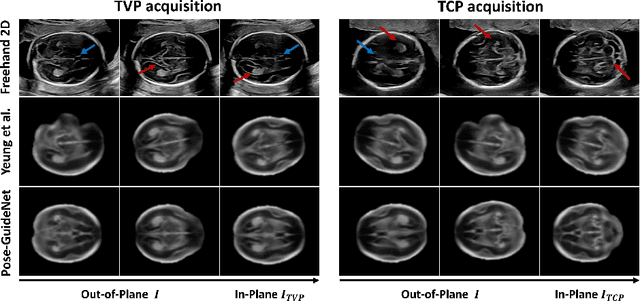
Abstract:3D pose estimation from a 2D cross-sectional view enables healthcare professionals to navigate through the 3D space, and such techniques initiate automatic guidance in many image-guided radiology applications. In this work, we investigate how estimating 3D fetal pose from freehand 2D ultrasound scanning can guide a sonographer to locate a head standard plane. Fetal head pose is estimated by the proposed Pose-GuideNet, a novel 2D/3D registration approach to align freehand 2D ultrasound to a 3D anatomical atlas without the acquisition of 3D ultrasound. To facilitate the 2D to 3D cross-dimensional projection, we exploit the prior knowledge in the atlas to align the standard plane frame in a freehand scan. A semantic-aware contrastive-based approach is further proposed to align the frames that are off standard planes based on their anatomical similarity. In the experiment, we enhance the existing assessment of freehand image localization by comparing the transformation of its estimated pose towards standard plane with the corresponding probe motion, which reflects the actual view change in 3D anatomy. Extensive results on two clinical head biometry tasks show that Pose-GuideNet not only accurately predicts pose but also successfully predicts the direction of the fetal head. Evaluations with probe motions further demonstrate the feasibility of adopting Pose-GuideNet for freehand ultrasound-assisted navigation in a sensor-free environment.
Dual Conditioned Diffusion Models for Out-Of-Distribution Detection: Application to Fetal Ultrasound Videos
Nov 01, 2023Abstract:Out-of-distribution (OOD) detection is essential to improve the reliability of machine learning models by detecting samples that do not belong to the training distribution. Detecting OOD samples effectively in certain tasks can pose a challenge because of the substantial heterogeneity within the in-distribution (ID), and the high structural similarity between ID and OOD classes. For instance, when detecting heart views in fetal ultrasound videos there is a high structural similarity between the heart and other anatomies such as the abdomen, and large in-distribution variance as a heart has 5 distinct views and structural variations within each view. To detect OOD samples in this context, the resulting model should generalise to the intra-anatomy variations while rejecting similar OOD samples. In this paper, we introduce dual-conditioned diffusion models (DCDM) where we condition the model on in-distribution class information and latent features of the input image for reconstruction-based OOD detection. This constrains the generative manifold of the model to generate images structurally and semantically similar to those within the in-distribution. The proposed model outperforms reference methods with a 12% improvement in accuracy, 22% higher precision, and an 8% better F1 score.
Show from Tell: Audio-Visual Modelling in Clinical Settings
Oct 25, 2023


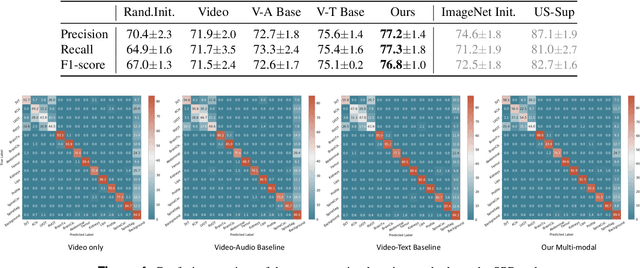
Abstract:Auditory and visual signals usually present together and correlate with each other, not only in natural environments but also in clinical settings. However, the audio-visual modelling in the latter case can be more challenging, due to the different sources of audio/video signals and the noise (both signal-level and semantic-level) in auditory signals -- usually speech. In this paper, we consider audio-visual modelling in a clinical setting, providing a solution to learn medical representations that benefit various clinical tasks, without human expert annotation. A simple yet effective multi-modal self-supervised learning framework is proposed for this purpose. The proposed approach is able to localise anatomical regions of interest during ultrasound imaging, with only speech audio as a reference. Experimental evaluations on a large-scale clinical multi-modal ultrasound video dataset show that the proposed self-supervised method learns good transferable anatomical representations that boost the performance of automated downstream clinical tasks, even outperforming fully-supervised solutions.
Anatomy-Aware Contrastive Representation Learning for Fetal Ultrasound
Aug 22, 2022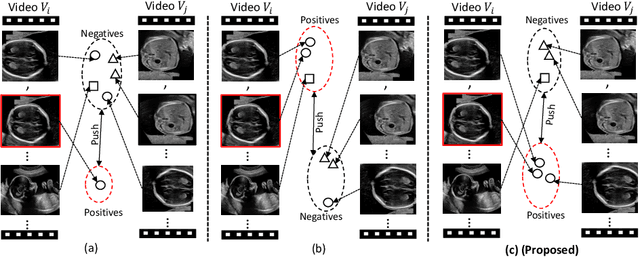

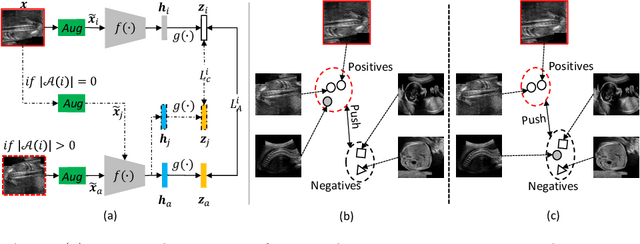
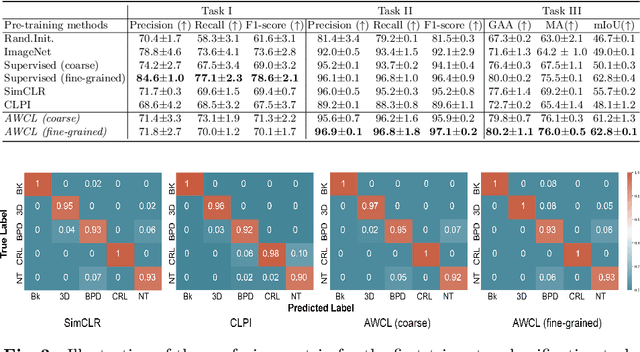
Abstract:Self-supervised contrastive representation learning offers the advantage of learning meaningful visual representations from unlabeled medical datasets for transfer learning. However, applying current contrastive learning approaches to medical data without considering its domain-specific anatomical characteristics may lead to visual representations that are inconsistent in appearance and semantics. In this paper, we propose to improve visual representations of medical images via anatomy-aware contrastive learning (AWCL), which incorporates anatomy information to augment the positive/negative pair sampling in a contrastive learning manner. The proposed approach is demonstrated for automated fetal ultrasound imaging tasks, enabling the positive pairs from the same or different ultrasound scans that are anatomically similar to be pulled together and thus improving the representation learning. We empirically investigate the effect of inclusion of anatomy information with coarse- and fine-grained granularity, for contrastive learning and find that learning with fine-grained anatomy information which preserves intra-class difference is more effective than its counterpart. We also analyze the impact of anatomy ratio on our AWCL framework and find that using more distinct but anatomically similar samples to compose positive pairs results in better quality representations. Experiments on a large-scale fetal ultrasound dataset demonstrate that our approach is effective for learning representations that transfer well to three clinical downstream tasks, and achieves superior performance compared to ImageNet supervised and the current state-of-the-art contrastive learning methods. In particular, AWCL outperforms ImageNet supervised method by 13.8% and state-of-the-art contrastive-based method by 7.1% on a cross-domain segmentation task.
Multimodal-GuideNet: Gaze-Probe Bidirectional Guidance in Obstetric Ultrasound Scanning
Jul 26, 2022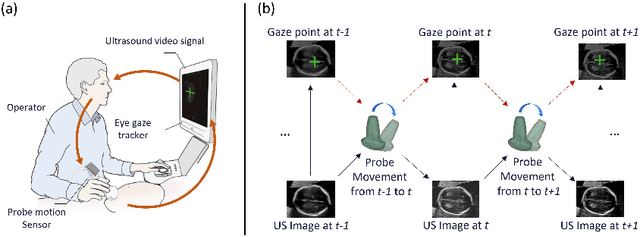
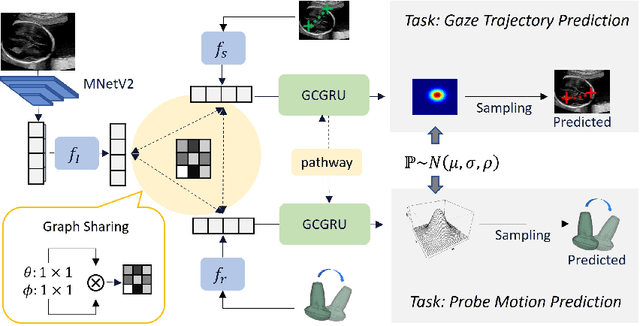
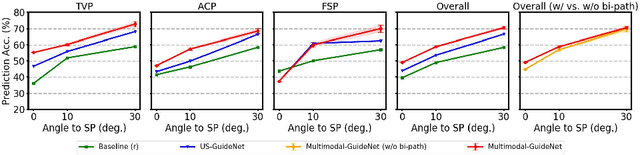
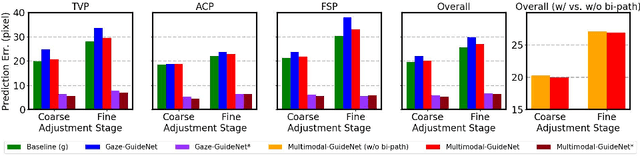
Abstract:Eye trackers can provide visual guidance to sonographers during ultrasound (US) scanning. Such guidance is potentially valuable for less experienced operators to improve their scanning skills on how to manipulate the probe to achieve the desired plane. In this paper, a multimodal guidance approach (Multimodal-GuideNet) is proposed to capture the stepwise dependency between a real-world US video signal, synchronized gaze, and probe motion within a unified framework. To understand the causal relationship between gaze movement and probe motion, our model exploits multitask learning to jointly learn two related tasks: predicting gaze movements and probe signals that an experienced sonographer would perform in routine obstetric scanning. The two tasks are associated by a modality-aware spatial graph to detect the co-occurrence among the multi-modality inputs and share useful cross-modal information. Instead of a deterministic scanning path, Multimodal-GuideNet allows for scanning diversity by estimating the probability distribution of real scans. Experiments performed with three typical obstetric scanning examinations show that the new approach outperforms single-task learning for both probe motion guidance and gaze movement prediction. Multimodal-GuideNet also provides a visual guidance signal with an error rate of less than 10 pixels for a 224x288 US image.
Self-Supervised Ultrasound to MRI Fetal Brain Image Synthesis
Aug 19, 2020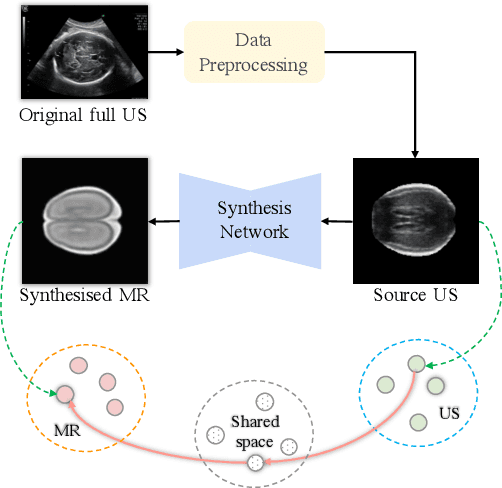
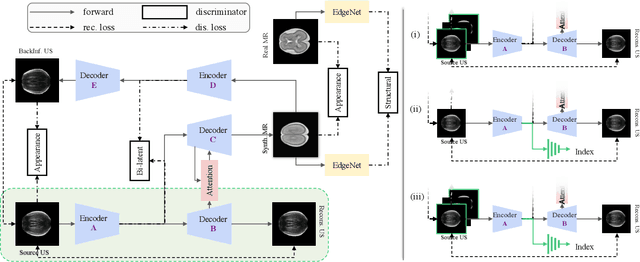
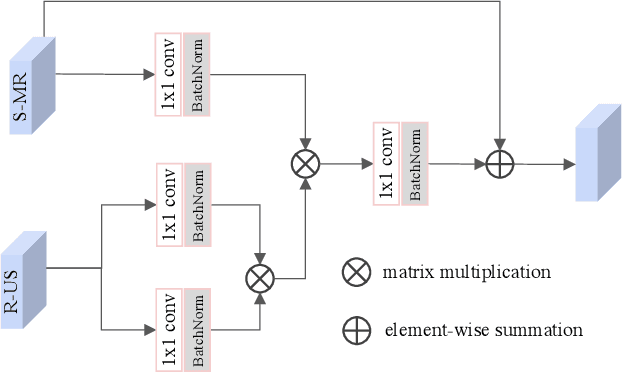
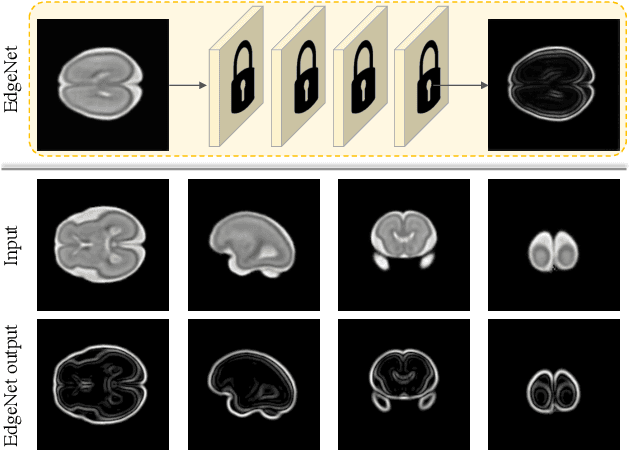
Abstract:Fetal brain magnetic resonance imaging (MRI) offers exquisite images of the developing brain but is not suitable for second-trimester anomaly screening, for which ultrasound (US) is employed. Although expert sonographers are adept at reading US images, MR images which closely resemble anatomical images are much easier for non-experts to interpret. Thus in this paper we propose to generate MR-like images directly from clinical US images. In medical image analysis such a capability is potentially useful as well, for instance for automatic US-MRI registration and fusion. The proposed model is end-to-end trainable and self-supervised without any external annotations. Specifically, based on an assumption that the US and MRI data share a similar anatomical latent space, we first utilise a network to extract the shared latent features, which are then used for MRI synthesis. Since paired data is unavailable for our study (and rare in practice), pixel-level constraints are infeasible to apply. We instead propose to enforce the distributions to be statistically indistinguishable, by adversarial learning in both the image domain and feature space. To regularise the anatomical structures between US and MRI during synthesis, we further propose an adversarial structural constraint. A new cross-modal attention technique is proposed to utilise non-local spatial information, by encouraging multi-modal knowledge fusion and propagation. We extend the approach to consider the case where 3D auxiliary information (e.g., 3D neighbours and a 3D location index) from volumetric data is also available, and show that this improves image synthesis. The proposed approach is evaluated quantitatively and qualitatively with comparison to real fetal MR images and other approaches to synthesis, demonstrating its feasibility of synthesising realistic MR images.
Self-supervised Contrastive Video-Speech Representation Learning for Ultrasound
Aug 14, 2020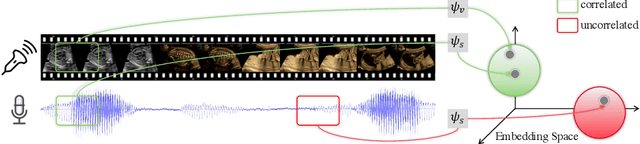


Abstract:In medical imaging, manual annotations can be expensive to acquire and sometimes infeasible to access, making conventional deep learning-based models difficult to scale. As a result, it would be beneficial if useful representations could be derived from raw data without the need for manual annotations. In this paper, we propose to address the problem of self-supervised representation learning with multi-modal ultrasound video-speech raw data. For this case, we assume that there is a high correlation between the ultrasound video and the corresponding narrative speech audio of the sonographer. In order to learn meaningful representations, the model needs to identify such correlation and at the same time understand the underlying anatomical features. We designed a framework to model the correspondence between video and audio without any kind of human annotations. Within this framework, we introduce cross-modal contrastive learning and an affinity-aware self-paced learning scheme to enhance correlation modelling. Experimental evaluations on multi-modal fetal ultrasound video and audio show that the proposed approach is able to learn strong representations and transfers well to downstream tasks of standard plane detection and eye-gaze prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge