Robail Yasrab
Anatomy-Aware Contrastive Representation Learning for Fetal Ultrasound
Aug 22, 2022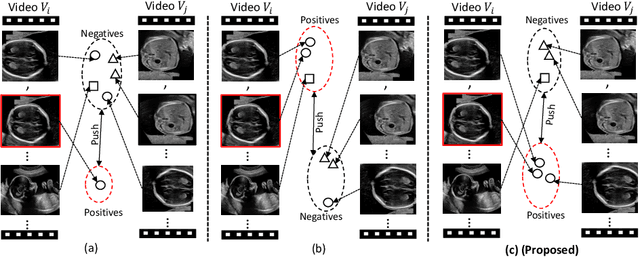

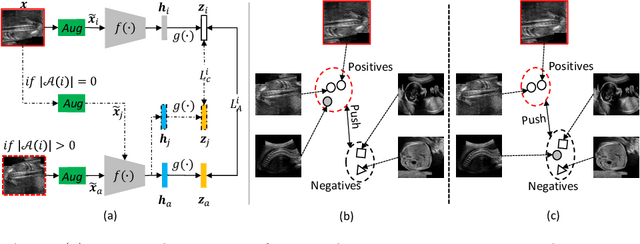
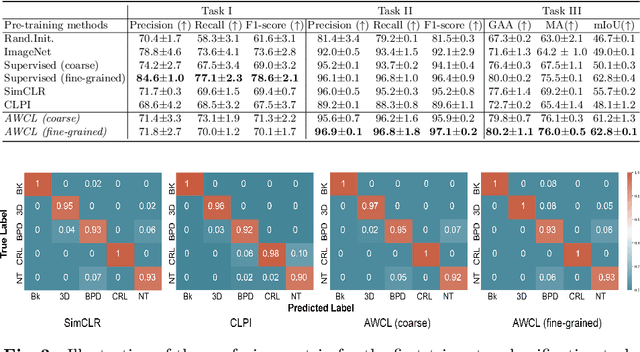
Abstract:Self-supervised contrastive representation learning offers the advantage of learning meaningful visual representations from unlabeled medical datasets for transfer learning. However, applying current contrastive learning approaches to medical data without considering its domain-specific anatomical characteristics may lead to visual representations that are inconsistent in appearance and semantics. In this paper, we propose to improve visual representations of medical images via anatomy-aware contrastive learning (AWCL), which incorporates anatomy information to augment the positive/negative pair sampling in a contrastive learning manner. The proposed approach is demonstrated for automated fetal ultrasound imaging tasks, enabling the positive pairs from the same or different ultrasound scans that are anatomically similar to be pulled together and thus improving the representation learning. We empirically investigate the effect of inclusion of anatomy information with coarse- and fine-grained granularity, for contrastive learning and find that learning with fine-grained anatomy information which preserves intra-class difference is more effective than its counterpart. We also analyze the impact of anatomy ratio on our AWCL framework and find that using more distinct but anatomically similar samples to compose positive pairs results in better quality representations. Experiments on a large-scale fetal ultrasound dataset demonstrate that our approach is effective for learning representations that transfer well to three clinical downstream tasks, and achieves superior performance compared to ImageNet supervised and the current state-of-the-art contrastive learning methods. In particular, AWCL outperforms ImageNet supervised method by 13.8% and state-of-the-art contrastive-based method by 7.1% on a cross-domain segmentation task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge