Nicola K Dinsdale
Automated quality assessment using appearance-based simulations and hippocampus segmentation on low-field paediatric brain MR images
Oct 08, 2024



Abstract:Understanding the structural growth of paediatric brains is a key step in the identification of various neuro-developmental disorders. However, our knowledge is limited by many factors, including the lack of automated image analysis tools, especially in Low and Middle Income Countries from the lack of high field MR images available. Low-field systems are being increasingly explored in these countries, and, therefore, there is a need to develop automated image analysis tools for these images. In this work, as a preliminary step, we consider two tasks: 1) automated quality assurance and 2) hippocampal segmentation, where we compare multiple approaches. For the automated quality assurance task a DenseNet combined with appearance-based transformations for synthesising artefacts produced the best performance, with a weighted accuracy of 82.3%. For the segmentation task, registration of an average atlas performed the best, with a final Dice score of 0.61. Our results show that although the images can provide understanding of large scale pathologies and gross scale anatomical development, there still remain barriers for their use for more granular analyses.
Geometric Transformation Uncertainty for Improving 3D Fetal Brain Pose Prediction from Freehand 2D Ultrasound Videos
May 21, 2024Abstract:Accurately localizing two-dimensional (2D) ultrasound (US) fetal brain images in the 3D brain, using minimal computational resources, is an important task for automated US analysis of fetal growth and development. We propose an uncertainty-aware deep learning model for automated 3D plane localization in 2D fetal brain images. Specifically, a multi-head network is trained to jointly regress 3D plane pose from 2D images in terms of different geometric transformations. The model explicitly learns to predict uncertainty to allocate higher weight to inputs with low variances across different transformations to improve performance. Our proposed method, QAERTS, demonstrates superior pose estimation accuracy than the state-of-the-art and most of the uncertainty-based approaches, leading to 9% improvement on plane angle (PA) for localization accuracy, and 8% on normalized cross-correlation (NCC) for sampled image quality. QAERTS also demonstrates efficiency, containing 5$\times$ fewer parameters than ensemble-based approach, making it advantageous in resource-constrained settings. In addition, QAERTS proves to be more robust to noise effects observed in freehand US scanning by leveraging rotational discontinuities and explicit output uncertainties.
SFHarmony: Source Free Domain Adaptation for Distributed Neuroimaging Analysis
Mar 28, 2023



Abstract:To represent the biological variability of clinical neuroimaging populations, it is vital to be able to combine data across scanners and studies. However, different MRI scanners produce images with different characteristics, resulting in a domain shift known as the `harmonisation problem'. Additionally, neuroimaging data is inherently personal in nature, leading to data privacy concerns when sharing the data. To overcome these barriers, we propose an Unsupervised Source-Free Domain Adaptation (SFDA) method, SFHarmony. Through modelling the imaging features as a Gaussian Mixture Model and minimising an adapted Bhattacharyya distance between the source and target features, we can create a model that performs well for the target data whilst having a shared feature representation across the data domains, without needing access to the source data for adaptation or target labels. We demonstrate the performance of our method on simulated and real domain shifts, showing that the approach is applicable to classification, segmentation and regression tasks, requiring no changes to the algorithm. Our method outperforms existing SFDA approaches across a range of realistic data scenarios, demonstrating the potential utility of our approach for MRI harmonisation and general SFDA problems. Our code is available at \url{https://github.com/nkdinsdale/SFHarmony}.
FedHarmony: Unlearning Scanner Bias with Distributed Data
May 31, 2022



Abstract:The ability to combine data across scanners and studies is vital for neuroimaging, to increase both statistical power and the representation of biological variability. However, combining datasets across sites leads to two challenges: first, an increase in undesirable non-biological variance due to scanner and acquisition differences - the harmonisation problem - and second, data privacy concerns due to the inherently personal nature of medical imaging data, meaning that sharing them across sites may risk violation of privacy laws. To overcome these restrictions, we propose FedHarmony: a harmonisation framework operating in the federated learning paradigm. We show that to remove the scanner-specific effects, we only need to share the mean and standard deviation of the learned features, helping to protect individual subjects' privacy. We demonstrate our approach across a range of realistic data scenarios, using real multi-site data from the ABIDE dataset, thus showing the potential utility of our method for MRI harmonisation across studies. Our code is available at https://github.com/nkdinsdale/FedHarmony.
Challenges for machine learning in clinical translation of big data imaging studies
Jul 07, 2021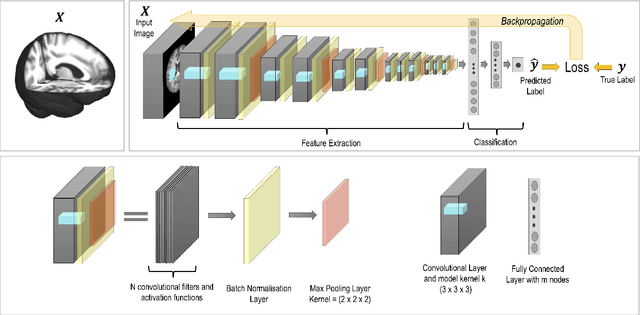
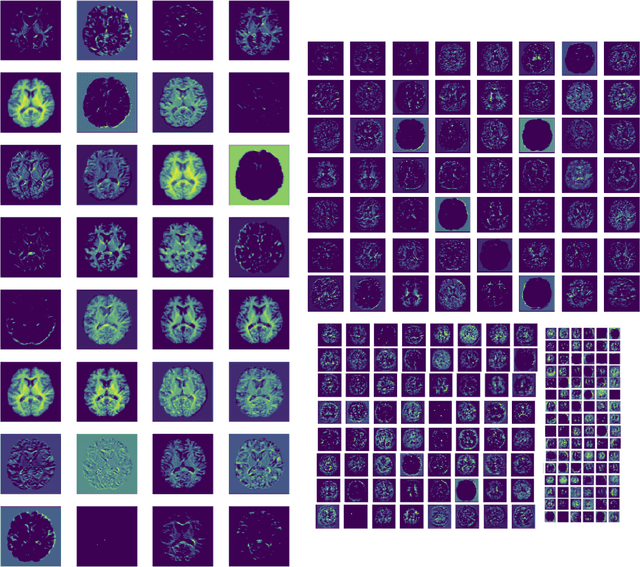
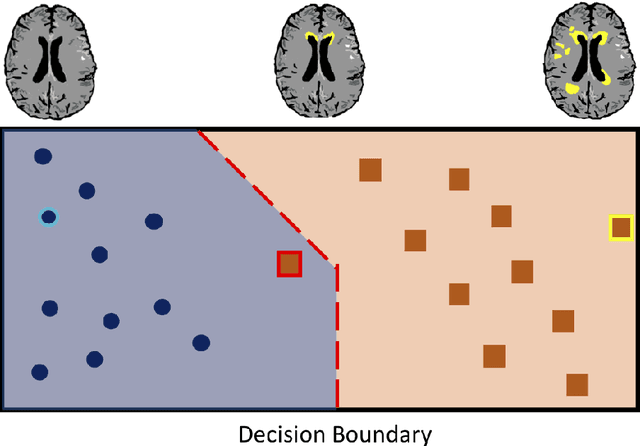
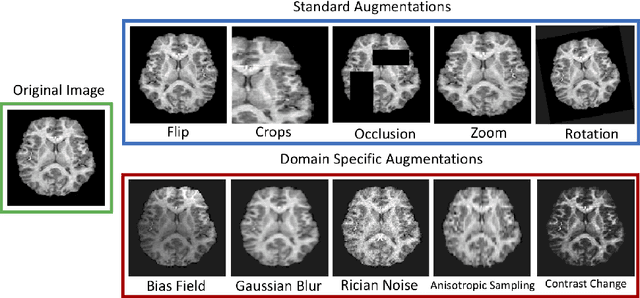
Abstract:The combination of deep learning image analysis methods and large-scale imaging datasets offers many opportunities to imaging neuroscience and epidemiology. However, despite the success of deep learning when applied to many neuroimaging tasks, there remain barriers to the clinical translation of large-scale datasets and processing tools. Here, we explore the main challenges and the approaches that have been explored to overcome them. We focus on issues relating to data availability, interpretability, evaluation and logistical challenges, and discuss the challenges we believe are still to be overcome to enable the full success of big data deep learning approaches to be experienced outside of the research field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge