Gabriel Maicas
Australian Institute for Machine Learning
Censor-aware Semi-supervised Learning for Survival Time Prediction from Medical Images
May 26, 2022



Abstract:Survival time prediction from medical images is important for treatment planning, where accurate estimations can improve healthcare quality. One issue affecting the training of survival models is censored data. Most of the current survival prediction approaches are based on Cox models that can deal with censored data, but their application scope is limited because they output a hazard function instead of a survival time. On the other hand, methods that predict survival time usually ignore censored data, resulting in an under-utilization of the training set. In this work, we propose a new training method that predicts survival time using all censored and uncensored data. We propose to treat censored data as samples with a lower-bound time to death and estimate pseudo labels to semi-supervise a censor-aware survival time regressor. We evaluate our method on pathology and x-ray images from the TCGA-GM and NLST datasets. Our results establish the state-of-the-art survival prediction accuracy on both datasets.
Mutual information neural estimation for unsupervised multi-modal registration of brain images
Jan 25, 2022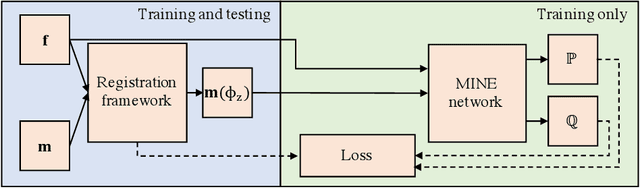
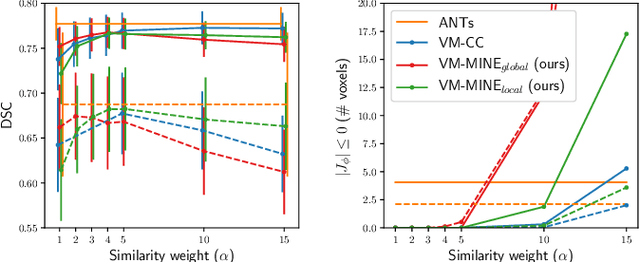
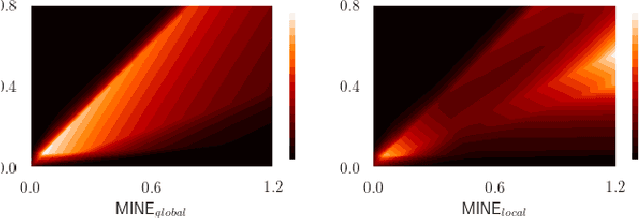

Abstract:Many applications in image-guided surgery and therapy require fast and reliable non-linear, multi-modal image registration. Recently proposed unsupervised deep learning-based registration methods have demonstrated superior performance compared to iterative methods in just a fraction of the time. Most of the learning-based methods have focused on mono-modal image registration. The extension to multi-modal registration depends on the use of an appropriate similarity function, such as the mutual information (MI). We propose guiding the training of a deep learning-based registration method with MI estimation between an image-pair in an end-to-end trainable network. Our results show that a small, 2-layer network produces competitive results in both mono- and multimodal registration, with sub-second run-times. Comparisons to both iterative and deep learning-based methods show that our MI-based method produces topologically and qualitatively superior results with an extremely low rate of non-diffeomorphic transformations. Real-time clinical application will benefit from a better visual matching of anatomical structures and less registration failures/outliers.
Post-hoc Overall Survival Time Prediction from Brain MRI
Feb 22, 2021



Abstract:Overall survival (OS) time prediction is one of the most common estimates of the prognosis of gliomas and is used to design an appropriate treatment planning. State-of-the-art (SOTA) methods for OS time prediction follow a pre-hoc approach that require computing the segmentation map of the glioma tumor sub-regions (necrotic, edema tumor, enhancing tumor) for estimating OS time. However, the training of the segmentation methods require ground truth segmentation labels which are tedious and expensive to obtain. Given that most of the large-scale data sets available from hospitals are unlikely to contain such precise segmentation, those SOTA methods have limited applicability. In this paper, we introduce a new post-hoc method for OS time prediction that does not require segmentation map annotation for training. Our model uses medical image and patient demographics (represented by age) as inputs to estimate the OS time and to estimate a saliency map that localizes the tumor as a way to explain the OS time prediction in a post-hoc manner. It is worth emphasizing that although our model can localize tumors, it uses only the ground truth OS time as training signal, i.e., no segmentation labels are needed. We evaluate our post-hoc method on the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2019 data set and show that it achieves competitive results compared to pre-hoc methods with the advantage of not requiring segmentation labels for training.
Detecting, Localising and Classifying Polyps from Colonoscopy Videos using Deep Learning
Jan 09, 2021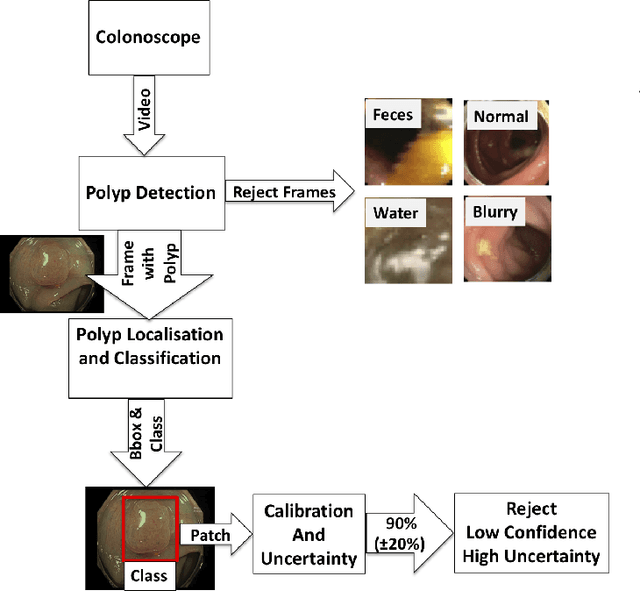
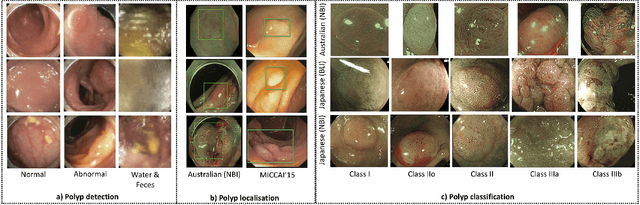
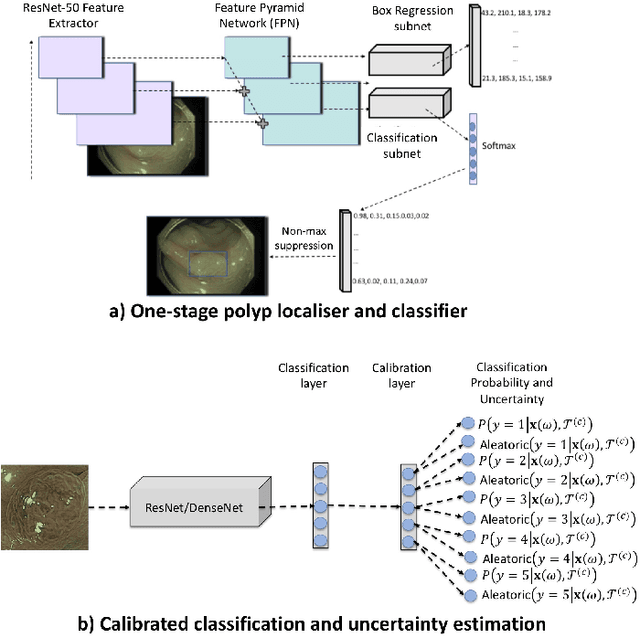
Abstract:In this paper, we propose and analyse a system that can automatically detect, localise and classify polyps from colonoscopy videos. The detection of frames with polyps is formulated as a few-shot anomaly classification problem, where the training set is highly imbalanced with the large majority of frames consisting of normal images and a small minority comprising frames with polyps. Colonoscopy videos may contain blurry images and frames displaying feces and water jet sprays to clean the colon -- such frames can mistakenly be detected as anomalies, so we have implemented a classifier to reject these two types of frames before polyp detection takes place. Next, given a frame containing a polyp, our method localises (with a bounding box around the polyp) and classifies it into five different classes. Furthermore, we study a method to improve the reliability and interpretability of the classification result using uncertainty estimation and classification calibration. Classification uncertainty and calibration not only help improve classification accuracy by rejecting low-confidence and high-uncertain results, but can be used by doctors to decide how to decide on the classification of a polyp. All the proposed detection, localisation and classification methods are tested using large data sets and compared with relevant baseline approaches.
Self-supervised Depth Estimation to Regularise Semantic Segmentation in Knee Arthroscopy
Jul 05, 2020

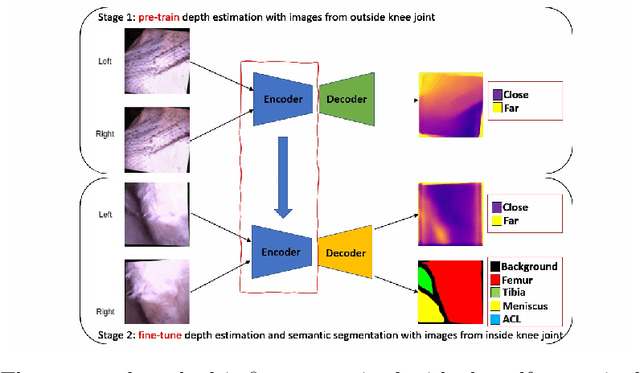
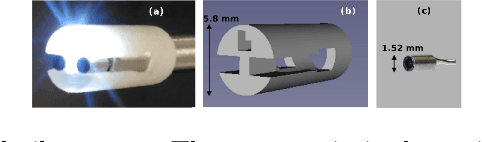
Abstract:Intra-operative automatic semantic segmentation of knee joint structures can assist surgeons during knee arthroscopy in terms of situational awareness. However, due to poor imaging conditions (e.g., low texture, overexposure, etc.), automatic semantic segmentation is a challenging scenario, which justifies the scarce literature on this topic. In this paper, we propose a novel self-supervised monocular depth estimation to regularise the training of the semantic segmentation in knee arthroscopy. To further regularise the depth estimation, we propose the use of clean training images captured by the stereo arthroscope of routine objects (presenting none of the poor imaging conditions and with rich texture information) to pre-train the model. We fine-tune such model to produce both the semantic segmentation and self-supervised monocular depth using stereo arthroscopic images taken from inside the knee. Using a data set containing 3868 arthroscopic images captured during cadaveric knee arthroscopy with semantic segmentation annotations, 2000 stereo image pairs of cadaveric knee arthroscopy, and 2150 stereo image pairs of routine objects, we show that our semantic segmentation regularised by self-supervised depth estimation produces a more accurate segmentation than a state-of-the-art semantic segmentation approach modeled exclusively with semantic segmentation annotation.
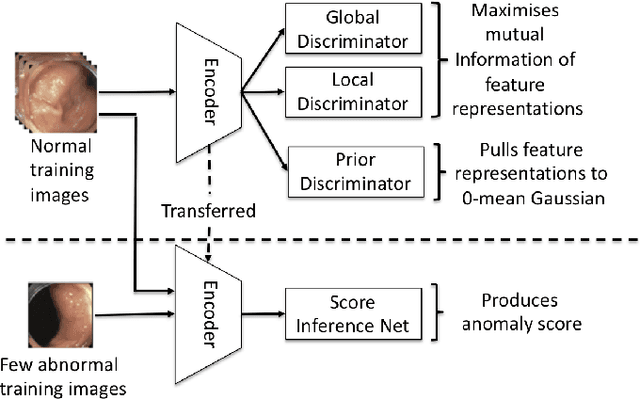
Few-Shot Anomaly Detection for Polyp Frames from Colonoscopy
Jun 26, 2020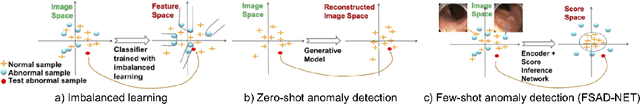
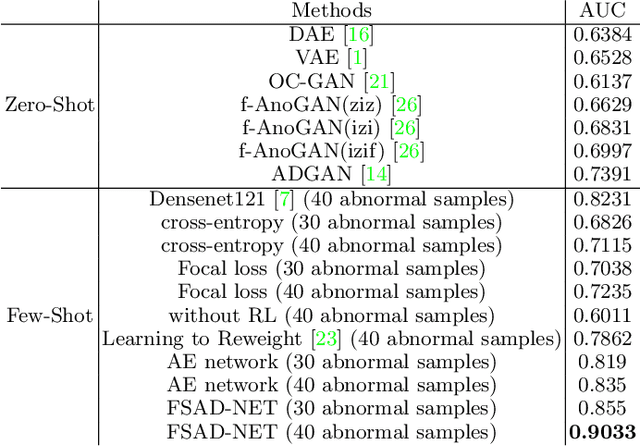
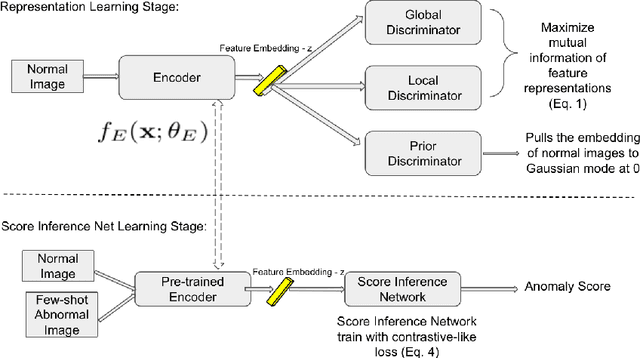
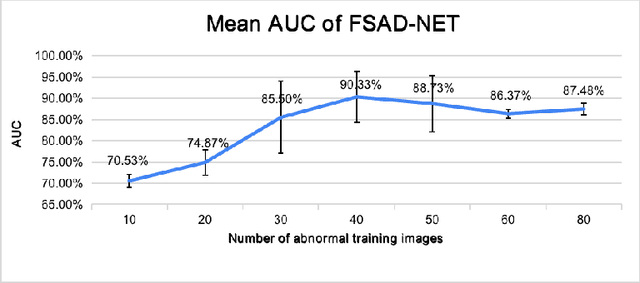
Abstract:Anomaly detection methods generally target the learning of a normal image distribution (i.e., inliers showing healthy cases) and during testing, samples relatively far from the learned distribution are classified as anomalies (i.e., outliers showing disease cases). These approaches tend to be sensitive to outliers that lie relatively close to inliers (e.g., a colonoscopy image with a small polyp). In this paper, we address the inappropriate sensitivity to outliers by also learning from inliers. We propose a new few-shot anomaly detection method based on an encoder trained to maximise the mutual information between feature embeddings and normal images, followed by a few-shot score inference network, trained with a large set of inliers and a substantially smaller set of outliers. We evaluate our proposed method on the clinical problem of detecting frames containing polyps from colonoscopy video sequences, where the training set has 13350 normal images (i.e., without polyps) and less than 100 abnormal images (i.e., with polyps). The results of our proposed model on this data set reveal a state-of-the-art detection result, while the performance based on different number of anomaly samples is relatively stable after approximately 40 abnormal training images.
Region Proposals for Saliency Map Refinement for Weakly-supervised Disease Localisation and Classification
May 22, 2020



Abstract:The deployment of automated systems to diagnose diseases from medical images is challenged by the requirement to localise the diagnosed diseases to justify or explain the classification decision. This requirement is hard to fulfil because most of the training sets available to develop these systems only contain global annotations, making the localisation of diseases a weakly supervised approach. The main methods designed for weakly supervised disease classification and localisation rely on saliency or attention maps that are not specifically trained for localisation, or on region proposals that can not be refined to produce accurate detections. In this paper, we introduce a new model that combines region proposal and saliency detection to overcome both limitations for weakly supervised disease classification and localisation. Using the ChestX-ray14 data set, we show that our proposed model establishes the new state-of-the-art for weakly-supervised disease diagnosis and localisation.
Semi-supervised Multi-domain Multi-task Training for Metastatic Colon Lymph Node Diagnosis From Abdominal CT
Oct 23, 2019


Abstract:The diagnosis of the presence of metastatic lymph nodes from abdominal computed tomography (CT) scans is an essential task performed by radiologists to guide radiation and chemotherapy treatment. State-of-the-art deep learning classifiers trained for this task usually rely on a training set containing CT volumes and their respective image-level (i.e., global) annotation. However, the lack of annotations for the localisation of the regions of interest (ROIs) containing lymph nodes can limit classification accuracy due to the small size of the relevant ROIs in this problem. The use of lymph node ROIs together with global annotations in a multi-task training process has the potential to improve classification accuracy, but the high cost involved in obtaining the ROI annotation for the same samples that have global annotations is a roadblock for this alternative. We address this limitation by introducing a new training strategy from two data sets: one containing the global annotations, and another (publicly available) containing only the lymph node ROI localisation. We term our new strategy semi-supervised multi-domain multi-task training, where the goal is to improve the diagnosis accuracy on the globally annotated data set by incorporating the ROI annotations from a different domain. Using a private data set containing global annotations and a public data set containing lymph node ROI localisation, we show that our proposed training mechanism improves the area under the ROC curve for the classification task compared to several training method baselines.
Photoshopping Colonoscopy Video Frames
Oct 23, 2019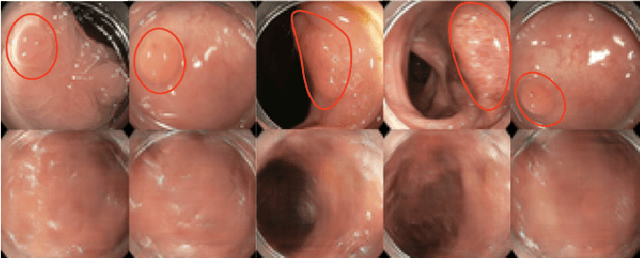
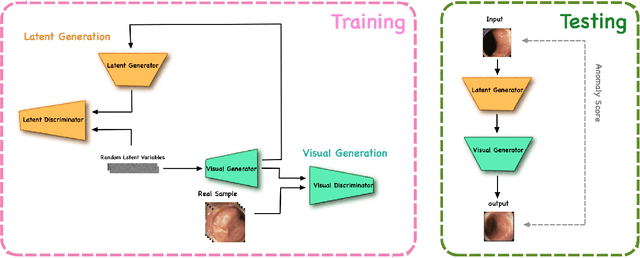
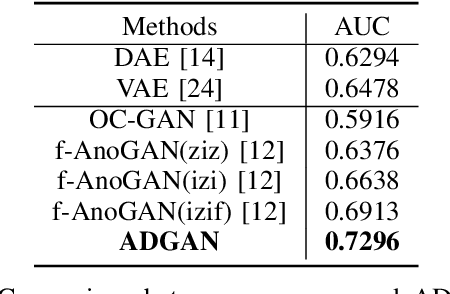
Abstract:The automatic detection of frames containing polyps from a colonoscopy video sequence is an important first step for a fully automated colonoscopy analysis tool. Typically, such detection system is built using a large annotated data set of frames with and without polyps, which is expensive to be obtained. In this paper, we introduce a new system that detects frames containing polyps as anomalies from a distribution of frames from exams that do not contain any polyps. The system is trained using a one-class training set consisting of colonoscopy frames without polyps -- such training set is considerably less expensive to obtain, compared to the 2-class data set mentioned above. During inference, the system is only able to reconstruct frames without polyps, and when it tries to reconstruct a frame with polyp, it automatically removes (i.e., photoshop) it from the frame -- the difference between the input and reconstructed frames is used to detect frames with polyps. We name our proposed model as anomaly detection generative adversarial network (ADGAN), comprising a dual GAN with two generators and two discriminators. We show that our proposed approach achieves the state-of-the-art result on this data set, compared with recently proposed anomaly detection systems.
Unsupervised Task Design to Meta-Train Medical Image Classifiers
Jul 17, 2019



Abstract:Meta-training has been empirically demonstrated to be the most effective pre-training method for few-shot learning of medical image classifiers (i.e., classifiers modeled with small training sets). However, the effectiveness of meta-training relies on the availability of a reasonable number of hand-designed classification tasks, which are costly to obtain, and consequently rarely available. In this paper, we propose a new method to unsupervisedly design a large number of classification tasks to meta-train medical image classifiers. We evaluate our method on a breast dynamically contrast enhanced magnetic resonance imaging (DCE-MRI) data set that has been used to benchmark few-shot training methods of medical image classifiers. Our results show that the proposed unsupervised task design to meta-train medical image classifiers builds a pre-trained model that, after fine-tuning, produces better classification results than other unsupervised and supervised pre-training methods, and competitive results with respect to meta-training that relies on hand-designed classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge